
And feel free to make suggestions on the article at the authors' Github repository: github.com/ispg-group/b...
29.01.2026 15:36 — 👍 4 🔁 1 💬 0 📌 0@livecomsjournal.bsky.social
The Living Journal of Computational Molecular Science (LiveCoMS) provides a peer-reviewed home for computational molecular science papers that can and should be updated. https://livecomsjournal.org/

And feel free to make suggestions on the article at the authors' Github repository: github.com/ispg-group/b...
29.01.2026 15:36 — 👍 4 🔁 1 💬 0 📌 0
The first article of volume 7 is out now!
Learn how to simulate molecular dynamics in electronic excited states, beyond the Born-Oppenheimer approximation, with this best practices article by Prlj et al on nonadiabatic dynamics! #compchem
doi.org/10.33011/liv...
As ever you are encouraged to make suggestions at the authors' Github repo: github.com/ManghraniA/M...
15.01.2026 08:58 — 👍 0 🔁 0 💬 0 📌 0
Interested in simulating modified nucleic acids?
The latest tutorial article by Galindo-Murillo et al details the steps needed to parameterize and run the simulations in the AMBER ecosystem with modXNA!
#compchem
doi.org/10.33011/liv...
As ever, you can comment on and contribute to the paper at the authors' GitHub repo: github.com/openkinome/k...
21.11.2025 14:50 — 👍 0 🔁 0 💬 0 📌 0
The latest article in our Lessons Learned category is out now!
"The Journey of Data: Lessons Learned in Modeling Kinase Affinity, Selectivity, and Resistance" by López-Ríos de Castro et al helps guide the development of platforms for structure-enabled ML for drug discovery: doi.org/10.33011/liv...
Chapin Cavendar's paper, Structure-Based Experimental Datasets for Benchmarking Protein Simulation Force Fields, is out now in LiveCoMS. Read it for a detailed look at the great work he has been doing toward an OpenFF protein force field, and stay tuned!
livecomsjournal.org/index.php/li...
Be sure to comment on the paper at the authors' Github repo: github.com/openforcefie...
28.10.2025 15:35 — 👍 1 🔁 0 💬 0 📌 0
In the latest @livecomsjournal.bsky.social
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem
Here's why we think this is still important today:
livecomsjournal.org/index.php/li...
But we'd love to continue that debate! What do you like about LiveCoMS, what made you publish with us or read an article? Or what can we do better, what is holding you off submitting your first manuscript?
Since then, we've published six volumes of peer-reviewed living documents, covering perpetually updated reviews, tutorials & software comparisons. Every article has its own GitHub repo for community discussion, and we're starting to publish 2nd versions with important scientific updates.
06.10.2025 15:21 — 👍 4 🔁 0 💬 1 📌 0
"Scientists are well aware that they seem to be getting a bad deal." www.theguardian.com/science/2017...
8 years ago, in response to this article, we wrote about the need for LiveCoMS, a diamond open access journal with no charge for submissions or for libraries: livecomsjournal.org/index.php/li...

You can find accompanying files and comment on the article here: github.com/lammpstutori...
02.10.2025 14:35 — 👍 0 🔁 0 💬 0 📌 0
📢 New software tutorial alert!
Gravelle et al introduce a suite of tutorials for new users of the LAMMPS simulation package, including molecular simulation basics, reactive force fields, GCMC and enhanced sampling #compchem
doi.org/10.33011/liv...
Hi, we have a few relevant tutorials out recently including MD with e.g. gromacs, gromos and biosimspace: livecomsjournal.org/index.php/li...
02.10.2025 06:12 — 👍 3 🔁 0 💬 0 📌 0
This is version 2 of this tutorial article, illustrating our 'living document' approach to publishing in which works are regularly updated on Github with community input. As ever, you can comment on this article here: github.com/biomos/gromo...
30.09.2025 17:06 — 👍 3 🔁 0 💬 0 📌 0
Interested in biomolecular simulation?
The latest article by Lier et al describes advanced tutorials for the GROMOS software, including free energy calculations, enhanced sampling and neural network potentials with SchNetPack! #compchem
livecomsjournal.org/index.php/li...
As ever you can comment on and contribute to the work via the accompanying GitHub repo: github.com/Foly93/MD_Fr...
09.09.2025 12:19 — 👍 4 🔁 0 💬 0 📌 0
The latest @livecomsjournal.bsky.social tutorial "Molecular Dynamics: From Basics to Application" by Vollmers, Chen et al is out now! doi.org/10.33011/liv...
It includes comprehensive MD tutorials in GROMACS, covering forcefields, thermodynamic ensembles, long-range electrostatics and much more!

Accompanying code is available at: github.com/liedllab/gis...
14.08.2025 16:44 — 👍 2 🔁 0 💬 0 📌 0
Interested in solvation free energies of small molecules and proteins?
The latest @livecomsjournal.bsky.social tutorial by Egger-Hoerschinger et al provides a guide to quantifying hydration thermodynamics using Grid Inhomogeneous Solvation Theory (GIST):
doi.org/10.33011/liv...
#compchem

You can comment on the manuscript and find code examples at the accompanying github repo: github.com/usnistgov/be...
11.08.2025 18:32 — 👍 2 🔁 0 💬 0 📌 0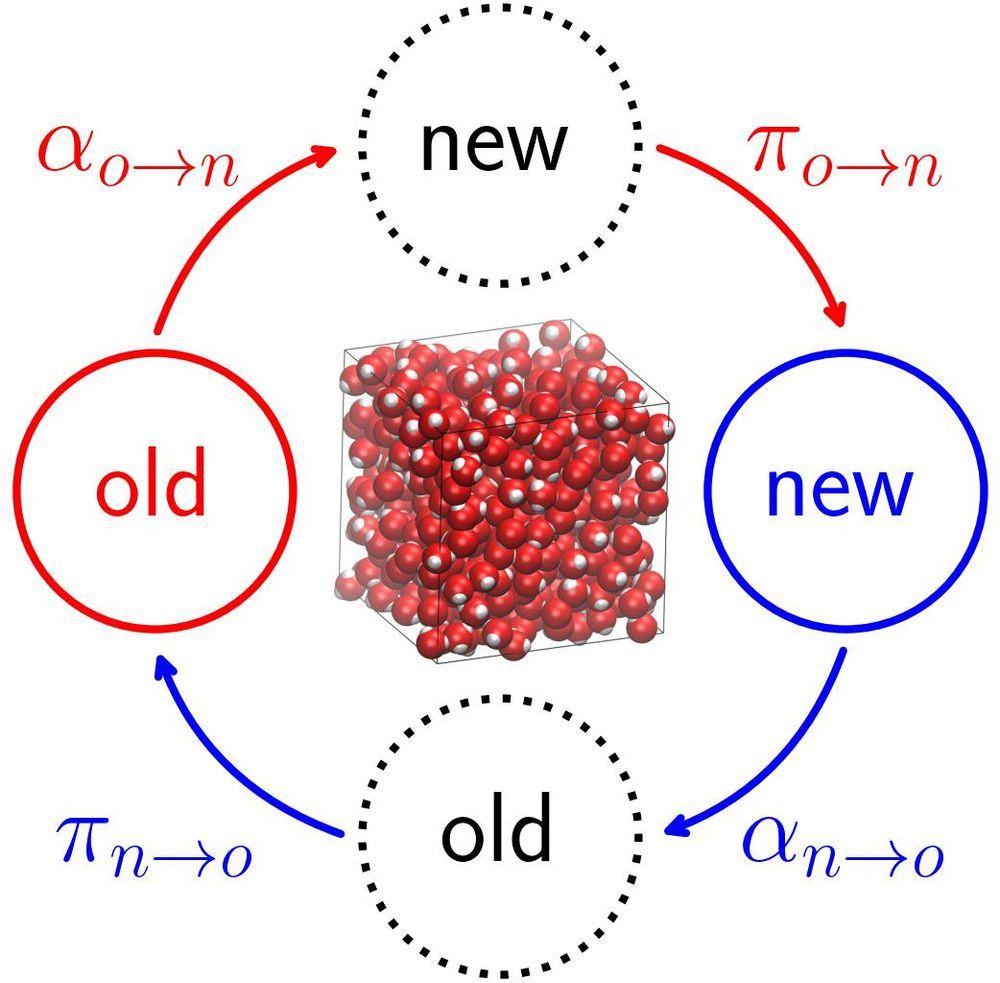
Table of contents figure
Our latest Best Practices article "Developing Monte Carlo Methodologies in Molecular Simulations" is out now and describes how to derive acceptance probabilities for a variety of Monte Carlo moves:
doi.org/10.33011/liv...
#compchem
As always, if you have any comments or questions on the paper, you're encouraged to engage with the paper's Github repository: github.com/Lemkul-Lab/g...
12.07.2025 19:55 — 👍 1 🔁 0 💬 0 📌 0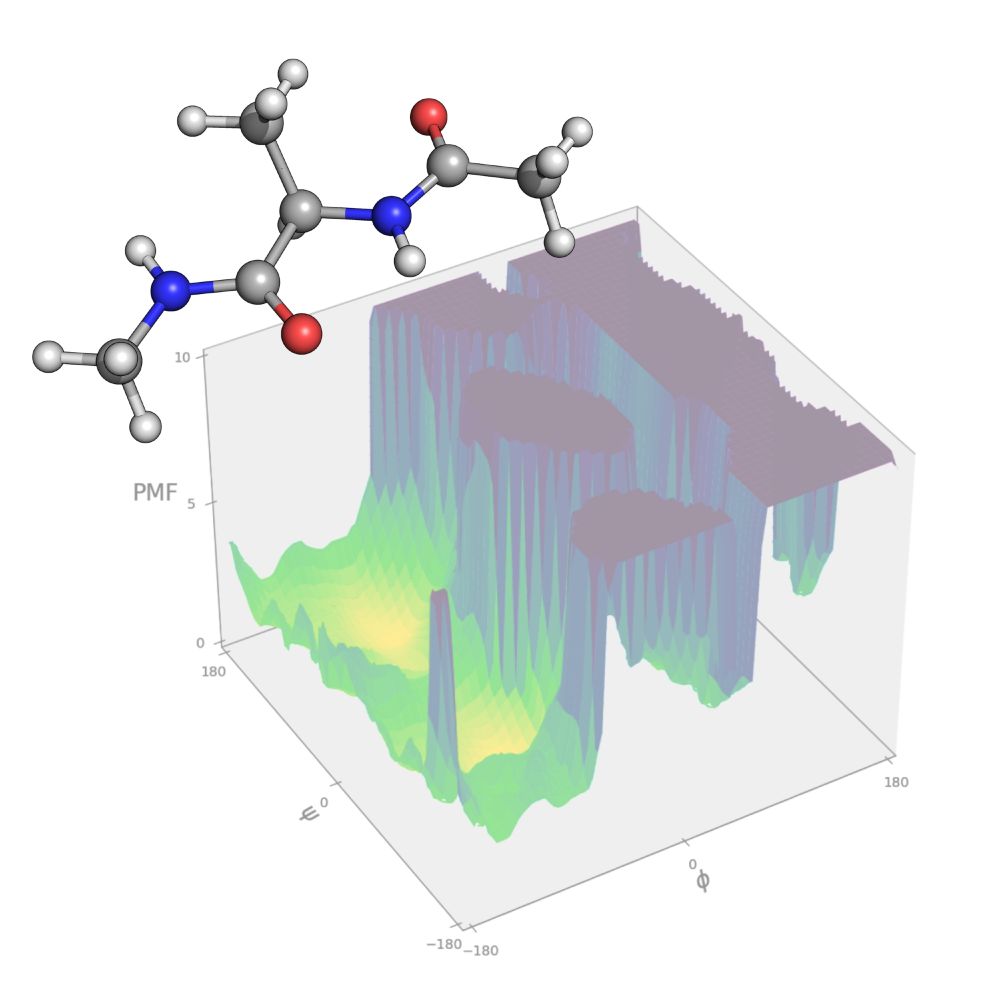
Interested in enhanced sampling for molecular dynamics simulations? Check out the latest LiveCoMS tutorial on Running Gaussian-accelerated Molecular Dynamics Simulations in NAMD:
livecomsjournal.org/index.php/li...
#compchem
Happy to announce our latest article, a tutorial on running Gaussian-accelerated MD simulations in NAMD. Tour de force work by @hmmichel.bsky.social with @mdpoleto.bsky.social that unifies theory and practice in one place!
livecomsjournal.org/index.php/li...
In the London heat, dreaming of a chemistry journal where articles evolve (living, versionable, open). Has anyone tried F1000? I am curious what the experience is like #OpenScience #ChemSky
21.06.2025 19:17 — 👍 12 🔁 1 💬 1 📌 0That's right - all articles are diamond open access and can be regularly updated!
21.06.2025 19:28 — 👍 4 🔁 0 💬 0 📌 0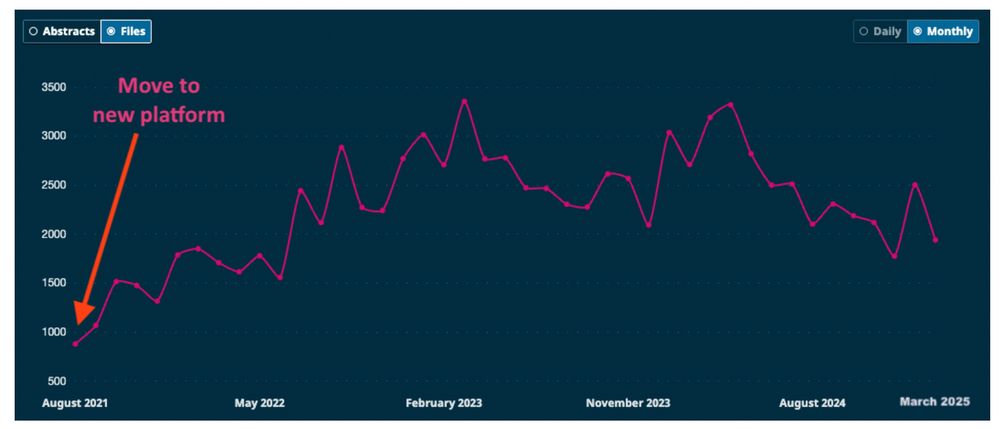
LiveCoMS monthly PDF downloads from our current hosting site, where we moved in late 2021
Do LiveCoMS articles make a mark? Check out our new blog post on our download & viewing statistics. We’re excited to see LiveCoMS is having an impact and large numbers of people find our work valuable! livecomsjournal.org/index.php/li...
25.03.2025 21:32 — 👍 4 🔁 2 💬 0 📌 0The Living Journal of Computational Molecular Science (LiveCoMS) is on BlueSky! Check out our recent blog post on why @livecomsjournal.bsky.social matters to find out more about us: livecomsjournal.org/index.php/li... #compchem #chemsky
30.01.2025 21:11 — 👍 21 🔁 9 💬 0 📌 0