
AML - outcome

ALL incidence
Finally out📯: 1st population-level picture of AML+ALL incidence & outcomes across all age groups in Germany! My colleague David Baden took the long tour to aggregate this data:
doi.org/10.1016/j.la... - Will be intro Figurę 1 to many presentations, David!🙏
20.10.2025 22:23 — 👍 2 🔁 0 💬 0 📌 0

Picture of Kiel University
1/4 🌍 We are looking for a passionate #PhD student in #Metabolic #Microbiome #Modeling in the Marie Skłodowska-Curie Doctoral Network MenoBrain. Join us at one of Europe’s microbiome research hubs and push the boundaries of microbiome science! @uni-kiel.de
🔗 www.uniklinikum-jena.de/menobrain/en...
09.10.2025 07:04 — 👍 10 🔁 9 💬 1 📌 3
Big shoutout to the whole group who made #IntegrateALL possible - especially @alinamh.bsky.social, Thomas Beder and Claudia Baldus at www.catchall-kfo5010.com
28.09.2025 08:36 — 👍 2 🔁 2 💬 0 📌 0
IntegrateALL: an end-to-end RNA-seq analysis pipeline for multilevel data extraction and interpretable subtype classification in B-precursor ALL https://www.biorxiv.org/content/10.1101/2025.09.25.673987v1
28.09.2025 03:49 — 👍 2 🔁 1 💬 0 📌 0

GitHub - NadineWolgast/IntegrateALL: Snakemake diagnostic Pipline for B-ALL classification
Snakemake diagnostic Pipline for B-ALL classification - NadineWolgast/IntegrateALL
🔗 Get the pipeline here:
github.com/NadineWolgas...
We welcome your feedback, issues & pull requests — and are happy to help you get started!🚀
#OpenSource #Bioinformatics #AcuteLymphoblasticLeukemia
28.09.2025 08:23 — 👍 4 🔁 0 💬 1 📌 1

In n=31/1210 B-ALL cases (2.6%), #IntegrateALL identified drivers from two different subtypes in one sample! One gene expression signature dominated in most cases, supporting unsupervised screening for drivers.
28.09.2025 08:23 — 👍 1 🔁 0 💬 1 📌 0

Analysis with our ICC/WHO-HAEM5 based rule-set yielded ~80% unambiguous subtype definitions based on driver calls and corresponding gene expression signatures, which was validated across independent cohorts. - Many thanks to collaborators @ www.mll.com!
28.09.2025 08:23 — 👍 0 🔁 0 💬 1 📌 0

With #IntegrateALL, we extract from RNA-seq:
- Expression counts
- B-ALL subtypes via #ALLCatchR
- Driver gene fusions
- SNVs
- Virtual karyotypes
For classifying #RNASeqCNV B-ALL karyotypes, we built #KaryALL, a machine-learning classifier reaching 0.98 accuracy!
28.09.2025 08:23 — 👍 0 🔁 0 💬 1 📌 0

Excited to share our preprint📯:
We introduce #IntegrateALL, an open-source RNA-seq pipeline for interpretable molecular diagnostics in acute lymphoblastic leukemia!🧬
Big thanks to Nadine Wolgast & team @ www.uksh.de
, catchall-kfo5010.com!
See: doi.org/p7k5 and 🧵
28.09.2025 08:23 — 👍 3 🔁 1 💬 1 📌 0
Finally some larger perspective after a long day on call… 🚀
07.08.2025 15:21 — 👍 1 🔁 0 💬 0 📌 0
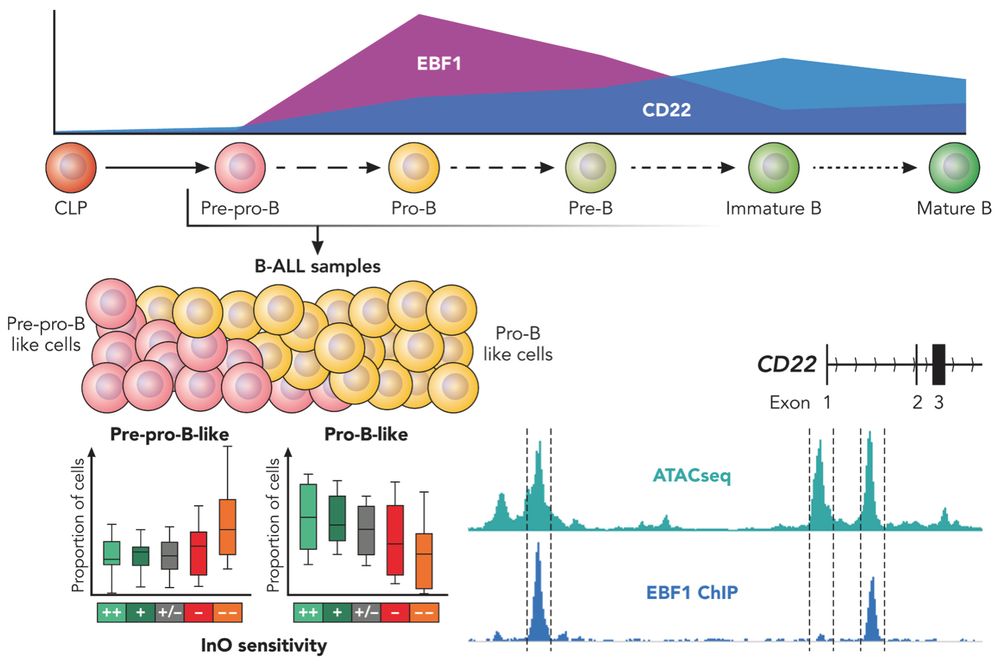
What determines Inotuzumab sensitivity in #ALL?
New @bloodjournal.bsky.social study from C. Escherich &
@junjyang.bsky.social Lab highlights the role of B cell developmental state and EBF1. It was a pleasure to provide a commentary!
🔬 doi.org/10.1182/bloo...
🗒️ doi.org/10.1182/bloo...
24.07.2025 21:09 — 👍 3 🔁 0 💬 0 📌 1
@Hemasphere_EHA acknowledged our ALLCatchR paper on machine learning based subtype classification in ALL as highly influential publication! 💥So happy and grateful! Big kudos to Thomas and our team at 👉catchall-kfo5010.com #EHA2025
20.06.2025 20:25 — 👍 4 🔁 0 💬 1 📌 0
So true!!
15.04.2025 18:47 — 👍 2 🔁 0 💬 0 📌 0
Mathematics Sorceror (sensory alchemist) at the Arctangent Transpetroglyphics Algra Laboratory (ATAL), I transflarnx mathematics into living rainbows. http://owen.maresh.info https://github.com/graveolensa
Psoeppe-Tlaxtlal, (an undreamt splendour?)
Assistant Professor at USC Center for Genetic Epidemiology. Researching the causes of childhood leukemia.
sites.usc.edu/childhoodcancer/
record.umn.edu
Clinician Scientist and Consultant Haematologist | Researches stem cell biology, epigenetics and drug discovery, specialising in B-ALL | Fellow St Catharine's College, Cambridge
Epigenetics enthusiast PDRA working on Super-enhancer hijacking in blood malignancies in the LEGEND group at Newcastle University
Head of Haematology/ Professor / Cancer and Leukaemia researcher / Cardiff University, School of Medicine / Supervisor / Educator / Collaborator
Cancer researcher at Hunter College of the City University of New York & Weill Cornell Medicine. Views are my own.
NUCancer | Newcastle University | Led by Prof. Anthony Moorman
https://research.ncl.ac.uk/lrcg/
A cross-border hub to develop & validate #AI techniques for anonymization & synthetic data generation in rare hematological diseases 🔬🩸🇪🇺 #HorizonEU project
👉 https://synthema.eu/
Understanding life. Advancing health.
Charité and Humboldt University Berlin, Computational Modelling, Cancer, Signaling
PhD candidate | Mathematical Modelling
European 🇪🇺 | #Berlin 🏠 | PhD student in medical systems biology 🧬 with @leifludwig.bsky.social | Cooking & mountains
Physics, Space, Quantum Computing | BTC: bc1qqu05pu3rdqjchnh37mtylrgprkk94udc3g3umj
https://thephysicsjournal.substack.com
He/Him
GitHub: https://github.com/rustcodepro
Bioinformatics Rust 🦀 Crates : https://crates.io/users/rustcodepro
Bioinformatician, Machine and Deep Learning. I write my code myself and ignore autocorrect typos. Read post on better ways in replies.
Kyushu University/hematology-oncology/B-cell precurser acute lympholastic leukemia/glucocorticoid/gymnastics/sake
Lab head in stem cell and cancer epigenetics at PeterMac, UniMelb 🇦🇺🧬
Scientist, mum, dreamer
She/her. Views are my own
www.eckmaslab.com
Drosophila lab based at Peter MacCallum Cancer Center, Melbourne, Australia. Interested in nutrition, cancer cachexia, neural development, environmental influences on organ size control.
Chenglabcom.wordpress.com
Immunologist & Group Leader at the Peter MacCallum Cancer Centre. Views are my own.
https://www.petermac.org/research/research-programs-and-labs/cancer-immunology-program/ian-parish-lab
Decoding the genome and unraveling molecular mysteries with precision and passion.
Cancer Metabolism Researcher | UNSW Sydney |
https://www.unsw.edu.au/staff/jeff-holst








