Elon Musk’s Worthless, Poisoned Hall of Mirrors
How X blew up its own platform with a new location feature
X’s decision to show where its accounts are based “is, theoretically, a positive step in the direction of transparency”—yet Elon Musk may have instead revealed that the platform is “just a worthless, poisoned hall of mirrors,” @cwarzel.bsky.social argues.
25.11.2025 07:45 — 👍 276 🔁 79 💬 11 📌 3

Three cichlid fish swimming in clear water, with text at the bottom asking 'How do 'supergenes' shape the evolution of fish?'
How did nature’s incredible diversity come to be? These cichlid fish may offer a clue 🐠
Cambridge researchers have found that chunks of ‘flipped’ #DNA can help fish quickly adapt to new habitats and evolve into new species, acting as evolutionary ‘superchargers’ 👇
bit.ly/3HGqEN6
13.06.2025 10:27 — 👍 19 🔁 10 💬 1 📌 3
And thanks to all other co-authors, some are on bluesky:
@currocam.bsky.social
@alexhooft.bsky.social
@joelelkin.bsky.social
@millanek.bsky.social
@dlimnothrissa.bsky.social
@domino-joyce.fishsci.com.ap.brid.gy
@astridboehne.bsky.social
@emiliapsantos.bsky.social
@ericmiska.bsky.social
19.06.2025 12:36 — 👍 1 🔁 0 💬 0 📌 0
A special thanks to everyone involved in this long-term project, especially the main authors Valentina Burskaia, Ilia Artiushin, @sahajaysmita.bsky.social, Hannes Svardal and Richard Durbin. Stay tuned for more interesting inversion stories!
19.06.2025 12:23 — 👍 0 🔁 1 💬 1 📌 0
Taken together, our findings point towards a dual role for recombination-suppressed regions (in our case: inversions) in the evolution of cichlid adaptive radiations. This is in line with another very nice recent study (Kumar et al., doi.org/10.7554/eLife.104923.2) who found the same inversions.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
(As a side note: We also found evidence that previously identified sex determination systems in the Lake Victoria sister radiation are most likely inversions, but emerged independent from the Malawi ones).
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Interestingly, we found strong evidence that at least three inversions are involved in sex determination in some lineages, while the same inversions do not determine sex in other groups.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Together with the origin of two inversions in the deepwater 𝐷𝑖𝑝𝑙𝑜𝑡𝑎𝑥𝑜𝑑𝑜𝑛 lineage, and the higher inversion frequencies in the extant deep clade, we believe that these inversions played a role when lineages adapted to different habitat depths, one of the most common axes of diversification in fish.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
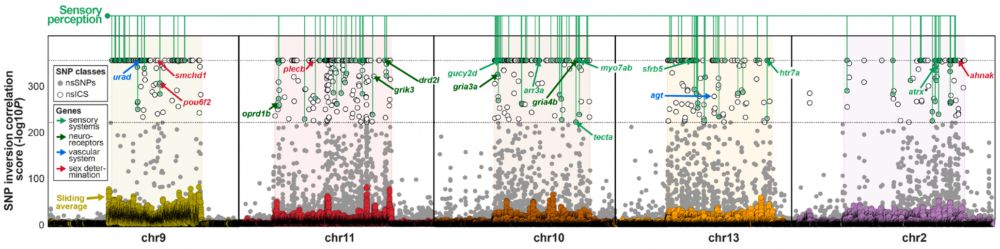
Focusing on the SNPs from within the inversion regions that were most correlated with inversion state, we identified patterns of positive selection that were enriched for genes related to sensory perception.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Interestingly, we found evidence that uninverted haplotypes of these inversions in benthics introgressed repeatedly from riverine lineages, in one case likely even from a 𝑃𝑠𝑒𝑢𝑑𝑜𝑐𝑟𝑒𝑛𝑖𝑙𝑎𝑏𝑟𝑢𝑠-like outgroup to the Malawi radiation, a lineage that still exists in the surrounding rivers today.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
In particular, the chromosome 9 and the compound chromosome 11 inversions likely arose in the proto 𝐷𝑖𝑝𝑙𝑜𝑡𝑎𝑥𝑜𝑑𝑜𝑛 lineage and remain polymorphic among today’s benthics.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
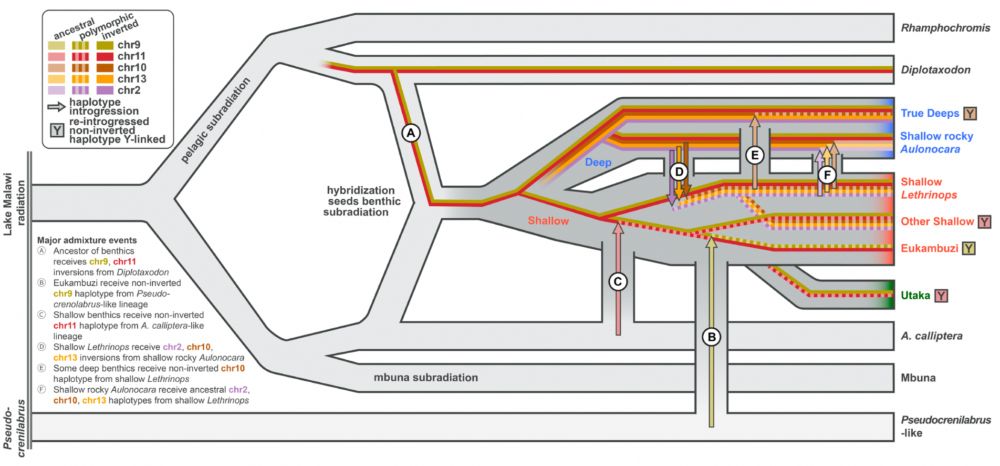
... we believe that the most species rich and ecologically diverse subradiation arose from a hybridization event between the ancestors of two ecologically contrasting lineages: the riverine-like 𝐴. 𝑐𝑎𝑙𝑙𝑖𝑝𝑡𝑒𝑟𝑎 and the deepwater specialists 𝐷𝑖𝑝𝑙𝑜𝑡𝑎𝑥𝑜𝑑𝑜𝑛.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Taking into account all available lines of evidence, especially the phylogenetic distribution of the inversions (see the first image) and patterns of allele sharing among Malawi clades and outgroups within and outside inversion regions ...
19.06.2025 12:23 — 👍 1 🔁 0 💬 1 📌 0
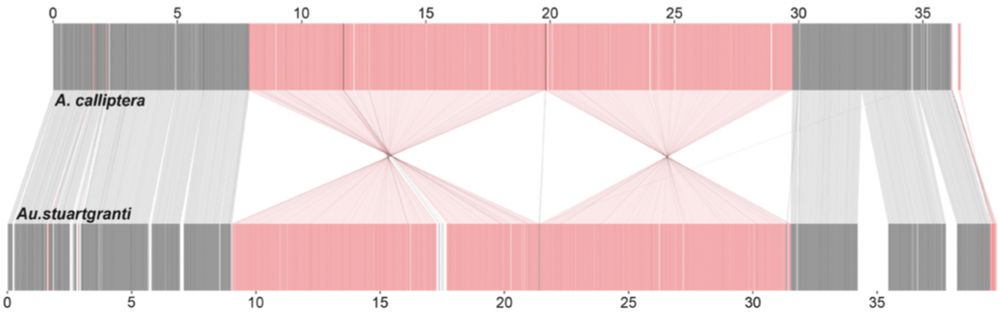
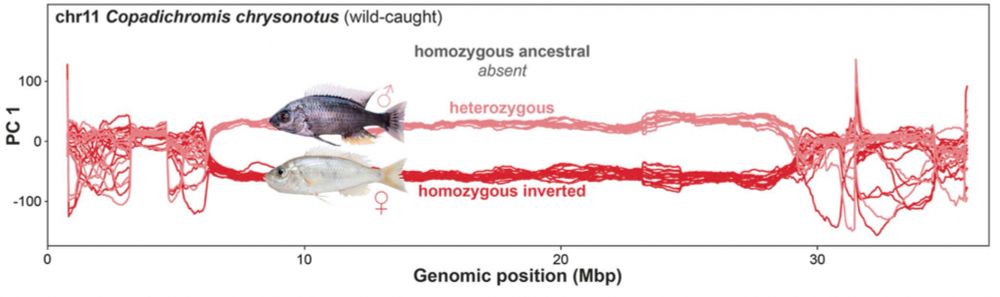
Using new chromosome-level assemblies and haplotagged (linked) reads of representatives of the major clades we confirmed that these regions are in fact inversions. Pairwise alignments also showed that the outlier region on chromosome 11 consists of two adjacent but co-segregating inversions.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
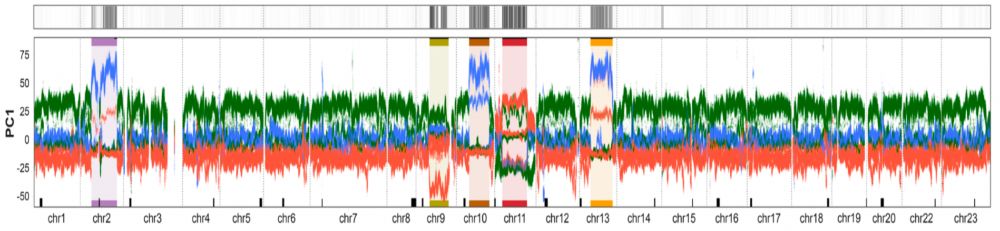
Using windowed PCA and a new clustering approach we identified five chromosome-scale regions with aberrant phylogenetic patterns – consistent with large and divergent segregating haplotypes.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Following a decade of field work with many collaborators from Europe and Malawi, we sequenced 1,375 Malawi cichlids from 240 species (covering all major ecomorphological clades) and called variants against a chromosome-level 𝐴. 𝑐𝑎𝑙𝑙𝑖𝑝𝑡𝑒𝑟𝑎 reference genome.
19.06.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
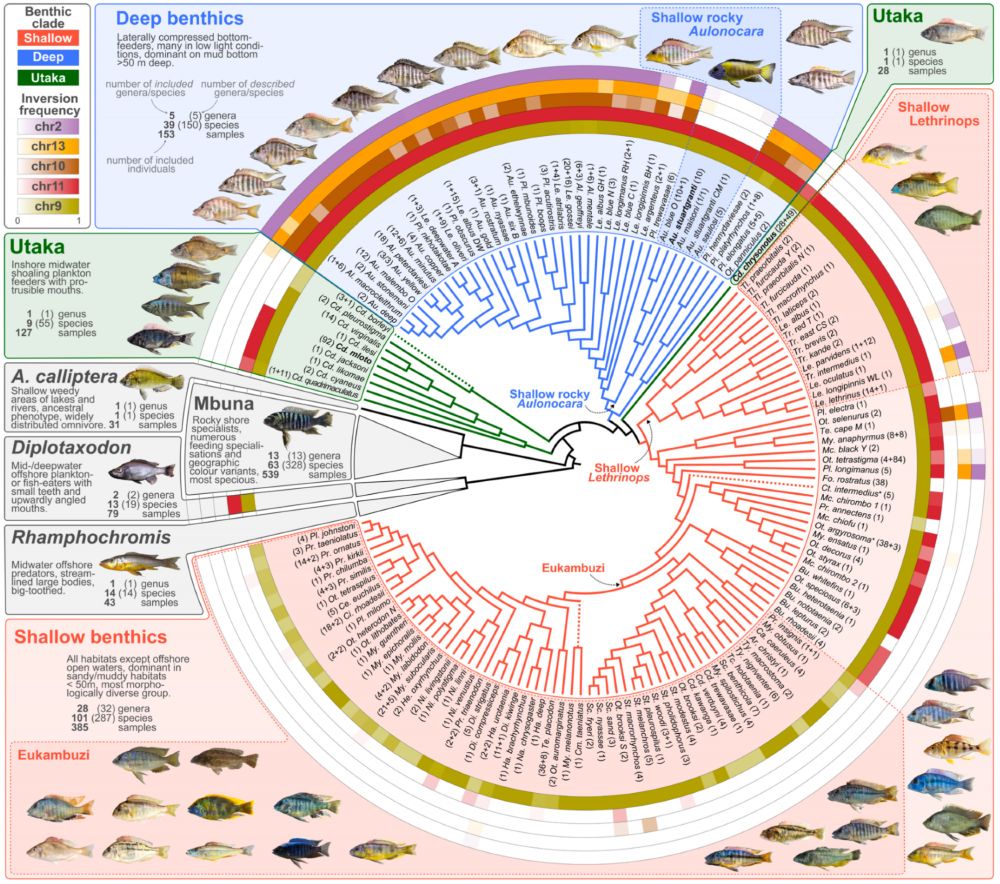
A circular phylogeny of Malawi cichlids with inversion frequencies indicated for different taxa as rings around the tree.
Check out our new paper about chromosomal inversions in Malawi cichlids! 🐟🧬
Available here without a paywall: hdl.handle.net/10067/214834... (click on the ‘Full text (open access)’ link).
19.06.2025 12:23 — 👍 19 🔁 11 💬 2 📌 1

Various Malawi cichlids in the shallow waters of Otter Point, Malawi
🐟 How do new #species arise?🧬 800+ species, 1 lake, little time: Cichlids in Lake #Malawi evolved with stunning speed—no geographic barriers needed. A new Science study shows how #supergenes & chromosome inversions drive biodiversity. #LIBresearch
Find out more: leibniz-lib.de/de/news/1206...
16.06.2025 09:07 — 👍 1 🔁 2 💬 0 📌 0

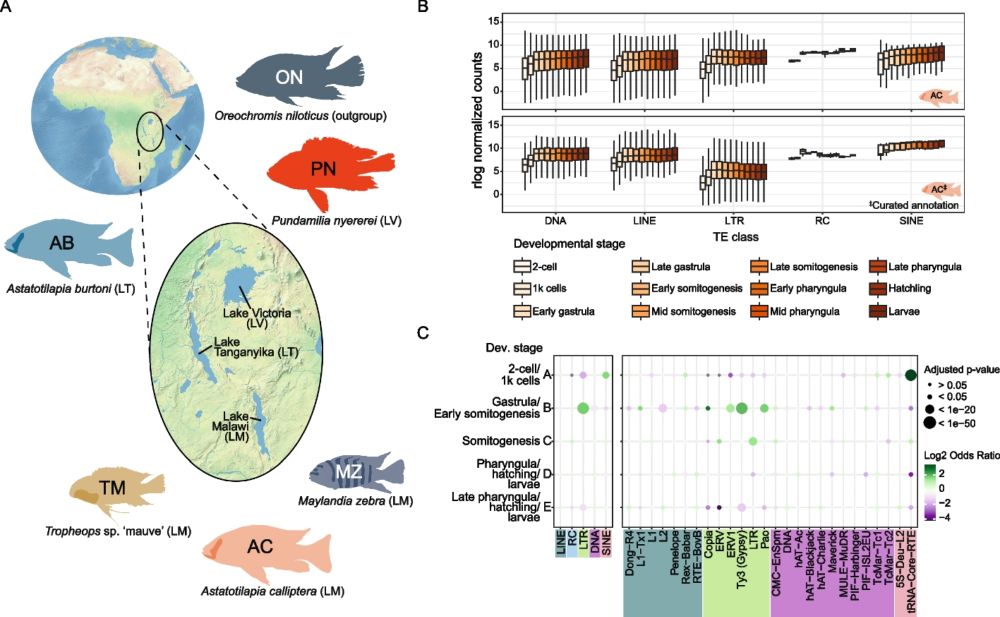
@fxquah.bsky.social 's paper is finally out @genomeresearch.bsky.social and we made the cover!! This was only possible due to the brilliant illustration by @sonhita.bsky.social
Paper here:
genome.cshlp.org/content/35/5...
See below for more details 🧵
06.05.2025 13:29 — 👍 35 🔁 12 💬 1 📌 3
Dass ausgerechnet die CSU-Politikerin Doro Bär, die noch vor wenigen Jahren den menschengemachten Klimawandel leugnete, jetzt neue Forschungsministerin werden soll - ein Job, bei dem es ganz besonders darauf ankommt, der Kraft der Wissenschaft zu vertrauen, ist m.E. hochproblematisch.
10.04.2025 14:52 — 👍 2633 🔁 683 💬 195 📌 62

Dire Wolves Were Not Really Wolves, New Genetic Clues Reveal
The extinct giant canids were a remarkable example of convergent evolution
Dire wolves were not close relatives of gray wolves. They last shared a common ancestor more than 5 million years ago. What Colossal has done is make something new and slapped a dire wolf sticker on it, as if an organism equals a hypothetical genome.
07.04.2025 14:56 — 👍 2595 🔁 883 💬 62 📌 128
Do you want to make genetic maps from sperm/pollen/gametes? Now there is an easy way, based on Hi-C sequencing.
Thanks to Richard Durbin and Ed Green for the idea and a group of co-authors including @mariontalbi.bsky.social and @danielbolnick.bsky.social for contributions.
12.03.2025 06:41 — 👍 49 🔁 22 💬 2 📌 0
Germans have been subjected to covert Russian propaganda as well as overt American propaganda - jointly in support of the far-right
www.nytimes.com/2025/02/13/w...
22.02.2025 17:13 — 👍 2934 🔁 1104 💬 75 📌 49
Postdoc at the University of Vienna.
The Integration of Speciation network aims to facilitate discussion and promote knowledge exchange between all corners of speciation research, including through our virtual seminar series. Find out more: https://speciation-network.pages.ist.ac.at/
Based in New Zealand. Research: #seafood #genomics #adaptation. Editor-In-Chief for @EvolAppJournal. Prof at @AucklandUni. Science Group Leader @@bioeconomyscience
https://marenwellenreuther.com/
Group leader, Senckenberg Research Institute, Frankfurt.
Former Sanger Institute, and Naturalis Biodiversity Center
Biodiversity #genomics, DNA-based monitoring of #biodiversity, #eDNA
Scientist Senckenberg Biodiversity and Climate Research Center, Frankfurt, Germany
evolutionary biology, adaptive genomics, behavior
Evolution, ecology & biological clocks - with a focus on lunar rhythms
in development & reproduction of the marine insect Clunio.
+ Genomics | Biodiversity | Behaviour | NeuroBio | MolBio | SciCom
bit.ly/KaiserLab
@mpi-evolbio.bsky.social @maxplanck.de
PhD student in Uppsala University and Zhejiang University
Science journalism by Leonid Schneider, on forbetterscience.com
Read at your own risk, people got sacked for much less. Posts may contain #antifa and #russophobia 🇺🇦
https://forbetterscience.com/
Dedicated to the study and conservation of the endangered Southern Resident Killer Whale (Orca) population in the Pacific Northwest since 1976
www.whaleresearch.com
Social Evolution, Life History Evolution, Conservation, Killer Whales. Prof of Behavioural Ecology at the University of Exeter https://experts.exeter.ac.uk/436-darren-croft - Executive Director of the Center for Whale Research https://www.whaleresearch.com
Website: https://www.geomar.de/en
Imprint: https://www.geomar.de/en/impressum
#OurWorldIsTheOcean
Assistant Professor @uarizona – Biological data science and macroevolution – Website: https://datadiversitylab.github.io/
Professor of ecology and evolution @ UConn. Studies adaptation, evolutionary immunology, speciation, genomics, foraging, parasite ecology and more.
Hikes, rock climbs, tango, history buff, photographer & parent
Dad, husband, and PhD 🧑🧑🧒🧒👨🏻🎓 Scientist, technologist, and engineer 👨🔬📱👨💻 Bibliophile 📚 Philomath 🤓 Mental health advocate 💚
Passionate about #bioinformatics, #computing, #evolution, #genomics, #medicine, and #sre 🧬🩺💻
Homepage: https://www.gawbul.io
Mexican Historian & Philosopher of Biology • Postdoctoral Fellow at @theramseylab.bsky.social (@clpskuleuven.bsky.social) • Book Reviews Editor for @jgps.bsky.social • https://www.alejandrofabregastejeda.com • #PhilSci #HistSTM #philsky • Escribo y edito
Evolutionary Biology | Ecology
I’m a biologist interested in the evolution of sex roles, animal behavior, and biodiversity conservation.
Undergrad in Biology (UFSCar - 🇧🇷)
Interested in evo-devo, genomics and speciation.
Embryo-stage evolutionary biologist.
Currently working with fruit-flies and adaptive radiations.
Welcome to the ESEB 2025 Congress, set to take place in Barcelona. This congress marks another milestone for the European Society for Evolutionary Biology (ESEB), which has been organising biennial conferences since 1987. Join us! We are waiting for you.
Colombian 🇨🇴 PhD student @ KU Leuven || Bioinformatician || Eco-evolutionary genomics 🪲🦋🕷️ Studying the evolutionary role of chromosomal inversions in beetles
🏳️⚧️ 🏳️🌈 🇵🇸