
Hominoid-specific retrotransposons fuel regulatory novelty in early brain development , by @retrogenomics.bsky.social.
➡️ www.cell.com/cell-genomic...
@manthan5277.bsky.social
Postdoc @Madapuralab, QMUL Applying Epigenomics Transposons NGS approaches understanding disease and development

Hominoid-specific retrotransposons fuel regulatory novelty in early brain development , by @retrogenomics.bsky.social.
➡️ www.cell.com/cell-genomic...

Many enhancers that drive tissue-specific gene expression are already connected to gene promoters in human pluripotent cells.
In a new preprint, we share some clues about when, how, and why this happens!
www.biorxiv.org/content/10.1...

Today in @nature.com, we present our work leveraging functional genomics and human blastoids to uncover a human-specific mechanism in preimplantation development driven by the endogenous retrovirus HERVK.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD

Histone acetylation - not always about transcriptional activation: H4K16ac safeguards the genome replication program by repressing premature replication of heterochromatin. Well done Marta Milan! tinyurl.com/yuy9m8t3
30.09.2025 12:14 — 👍 31 🔁 13 💬 2 📌 0
Emergence of activation or repression in transcriptional control under a fixed molecular context www.pnas.org/doi/10.1073/...
24.09.2025 09:22 — 👍 27 🔁 8 💬 0 📌 1Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇

Hepatic acetyl-CoA metabolism modulates neuroinflammation and depression susceptibility via acetate @cp-cellmetabolism.bsky.social
www.cell.com/cell-metabol...
Our study on the role of LTR5HS and SVAs in regulation of human neural crest migration is now published as peer-reviewed paper on @molsystbiol.org!
Congrats to first author brilliant postdoc Laura Deelen, and all the authors involved! @imperialsci.bsky.social
www.embopress.org/doi/full/10....

Check out our pre-print: We show that L1 insertion intermediates can recombine with one another or with DNA breaks to form genome rearrangements, highlighting the risk of L1 retrotransposition generating substrates for aberrant recombination and genome instability.
www.biorxiv.org/content/10.1...

✨Exciting news: the main story of my PhD is out in Science!
Together with Christine Moene @cmoene.bsky.social, we explored what happens when you scramble the genome—revealing how Sox2’s position shapes enhancer activation.
📖 Read the full story here: www.science.org/doi/10.1126/...
WDR5 and Myc Cooperate to Regulate Formation of Neural Crest Stem Cells https://www.biorxiv.org/content/10.1101/2025.09.19.677424v1
20.09.2025 08:30 — 👍 0 🔁 3 💬 0 📌 1Have you ever wondered how the exact location of a gene affects it's activity?
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you

Transposable elements are vectors of recurrent transgenerational epigenetic inheritance @science.org @pierrebaduel.bsky.social
www.science.org/doi/10.1126/...
Excited for a major milestone in our efforts to map enhancers and interpret variants in the human genome:
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/

ASOG: AntiSense Oligonucleotide Generator. #Oligonucleotides #ComputationalStructuralBiotechnology 🧬 🖥️
www.sciencedirect.com/science/arti...
Thrilled to share our story in its final form! nature.com/articles/s41... 💥 After ~10 years, we show that BMAL1 represses transposons via chromatin regulation in embryonic stem cells—rather than circadian rhythms as in adult tissues.
10.09.2025 14:26 — 👍 16 🔁 6 💬 1 📌 1Very proud of our paper on "scrambling-by-hopping" LADs, which was just published: www.nature.com/articles/s41.... Congrats to Lise Dauban and the rest of the team – this was a real tour-de-force!
02.09.2025 17:26 — 👍 57 🔁 24 💬 0 📌 0
"We propose a model in which TF binding is not determined by individual binding sites, but rather by the sum of multiple, overlapping binding sites."
www.nature.com/articles/s41...

New paper from Bin Cao, showing that placenta-specific KO of ADAR1 is embryonic lethal.
22.08.2025 11:28 — 👍 15 🔁 7 💬 0 📌 0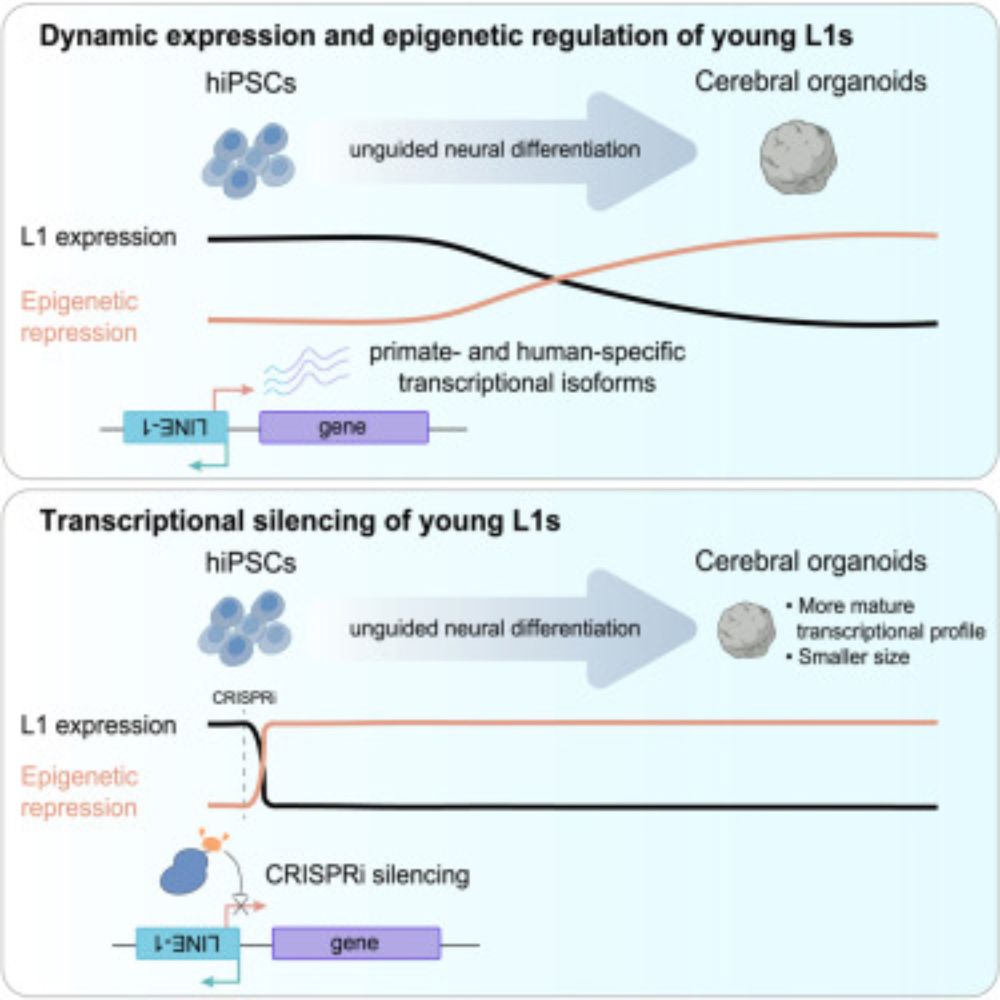
New paper from our lab! We found that LINE-1 transposons contribute to early human brain development.
Funded by @asapresearch.parkinsonsroadmap.org
www.cell.com/cell-genomic...

Excited to share our work on the specificity of PRC2 subcomplexes in stem cell and differentiation. We show that PRC2.1 and PRC2.2 exhibit distinct H3K27 methylation specificity in human pluripotent stem cells and opposing role in differentiation. Now online at Mol Cell: www.cell.com/molecular-ce...
12.08.2025 15:38 — 👍 30 🔁 10 💬 2 📌 0
Widespread 3′ UTR splicing regulates expression of oncogene transcripts through multiple mechanisms. #3UTRsplicing #TranscriptRegulation #Genomics @narjournal.bsky.social
academic.oup.com/nar/article/...

🚨New paper just dropped!🚨
We found that transposable element transcripts that are elevated in the blood of older adults and those with dementia are also epigenetically dysregulated, and may predict peripheral markers of inflammation & neurodegeneration
link.springer.com/article/10.1...

Really beautiful work led by Jessica Leismann in Joan Barau’s lab @imbmainz.bsky.social . If you like elegant mouse genetics, the transposon arms race, and a healthy dose of epigenetics, this one’s for you. www.embopress.org/doi/full/10.... @emboreports.org
06.08.2025 19:48 — 👍 51 🔁 23 💬 1 📌 1
Gene-Set Enrichment Analysis and visualization on the web using EnrichmentMap: RNASeq. #GSEA #WebTool #RNAseq #Bioinformatics @bioinfoadv.bsky.social 🧬 🖥️
academic.oup.com/bioinformati...
My first first author manuscript has posted! I’m very excited to share SWIF-TE with yall. It is a fast, memory efficient tool to identify novel TE insertions from short-read data. 🧬 #TEworldwide #transposons
02.08.2025 18:42 — 👍 77 🔁 19 💬 3 📌 3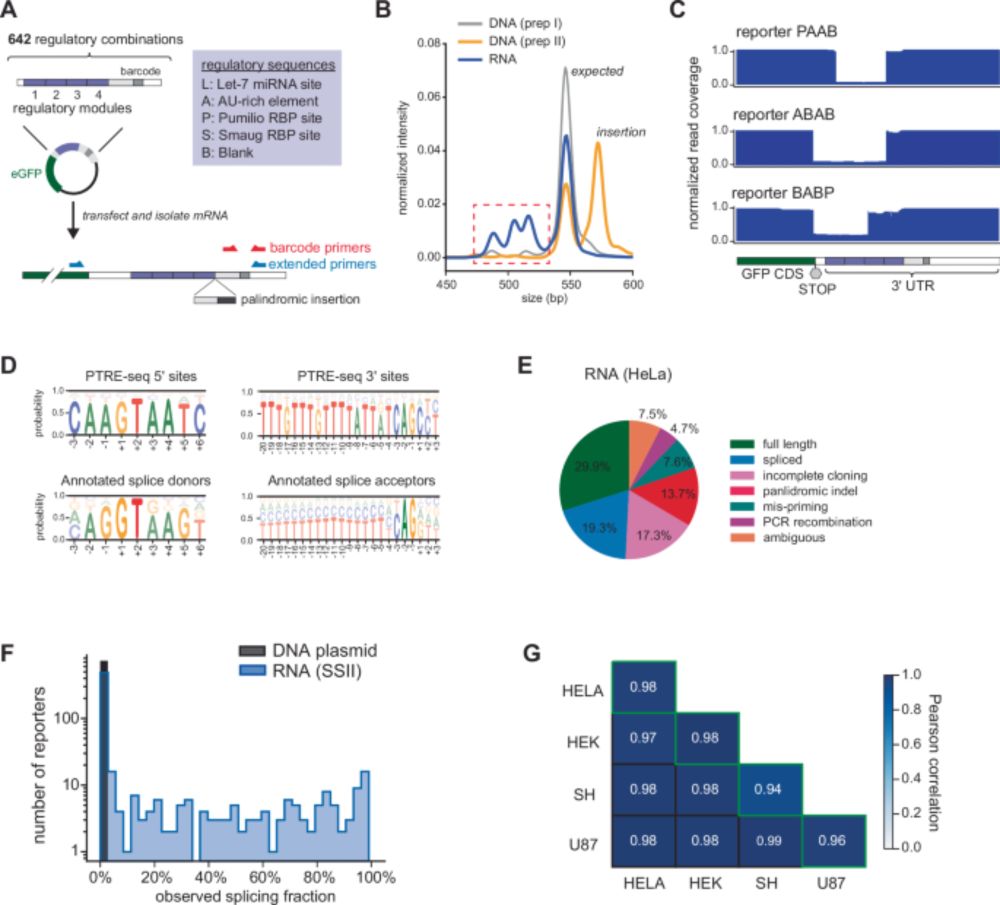
Our collaboration with Tony Mustoe's lab us out today. MPRA libraries and possible cryptic splicing in MPRA reporters, or reporters in general. The additional novelty here is that such events are controlled or influenced by AU-rich sequences.
www.nature.com/articles/s41...
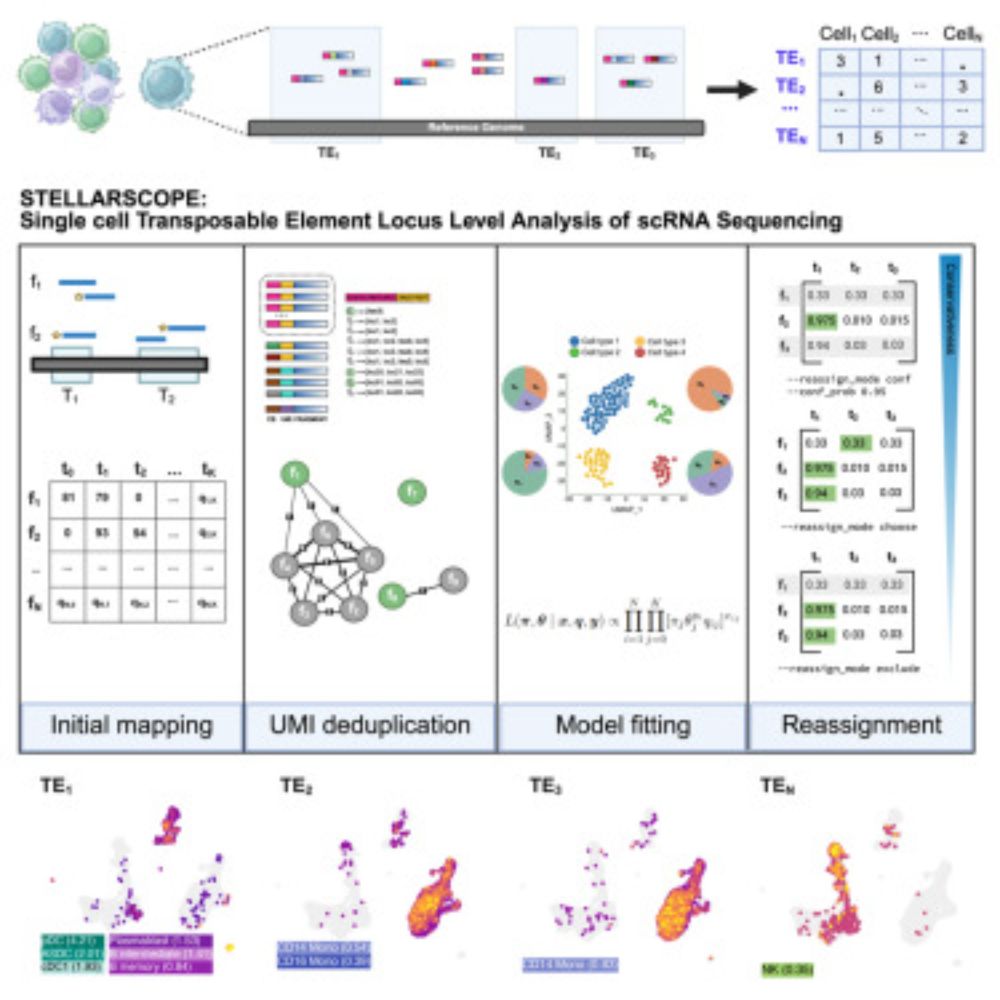
🧵1/ Introducing Stellarscope: an EM-based tool for quantifying locus-specific transposable element expression in single cells.
A single-cell transposable element atlas of human cell identity: Cell Reports Methods www.cell.com/cell-reports...
@hamarillo.bsky.social #TEHub #singlecell #genomics



Happy to share our recent publication on benchmarking peak calling methods for CUT&RUN.
academic.oup.com/bioinformati...
Published in @oxunipress.bsky.social Bioinformatics | led by aminnoorani.bsky.social
#Epigenomics #Genomics #Bioinformatics #MachineLearning #CUTandRUN #CompBio #4DNucleome