Proteins are at least a known thing with a standard code. I dont know if Metabolites even truly exist 👀 haha
Don't need some unknown unknowns 🤣
26.02.2025 01:53 — 👍 0 🔁 0 💬 0 📌 0
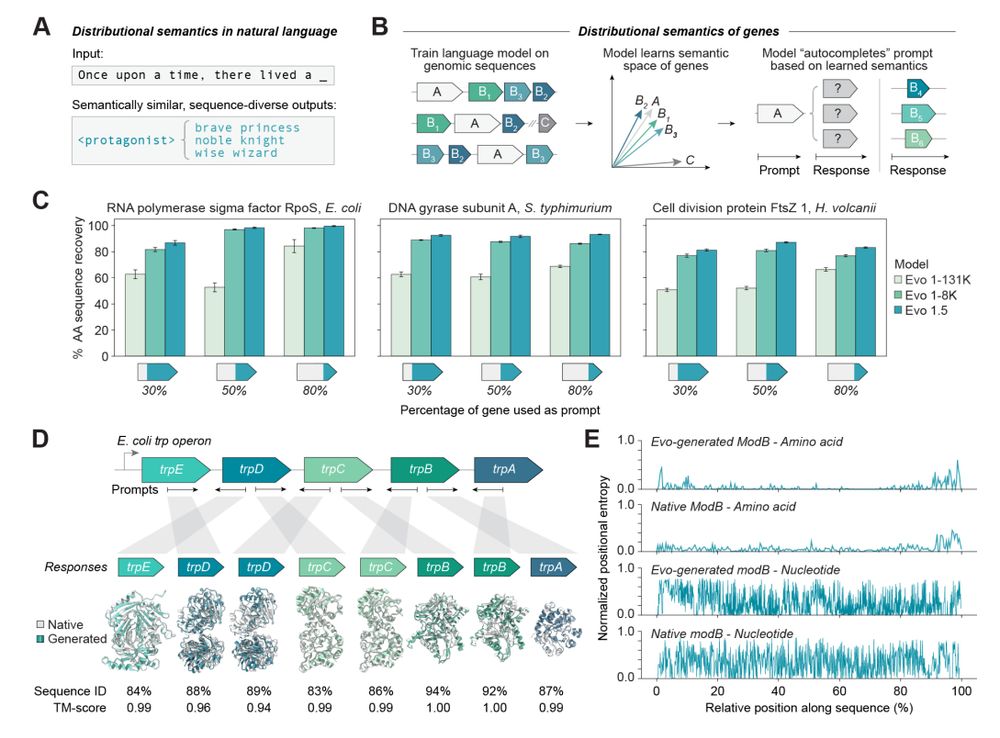
In-context genomic modeling and design with Evo.
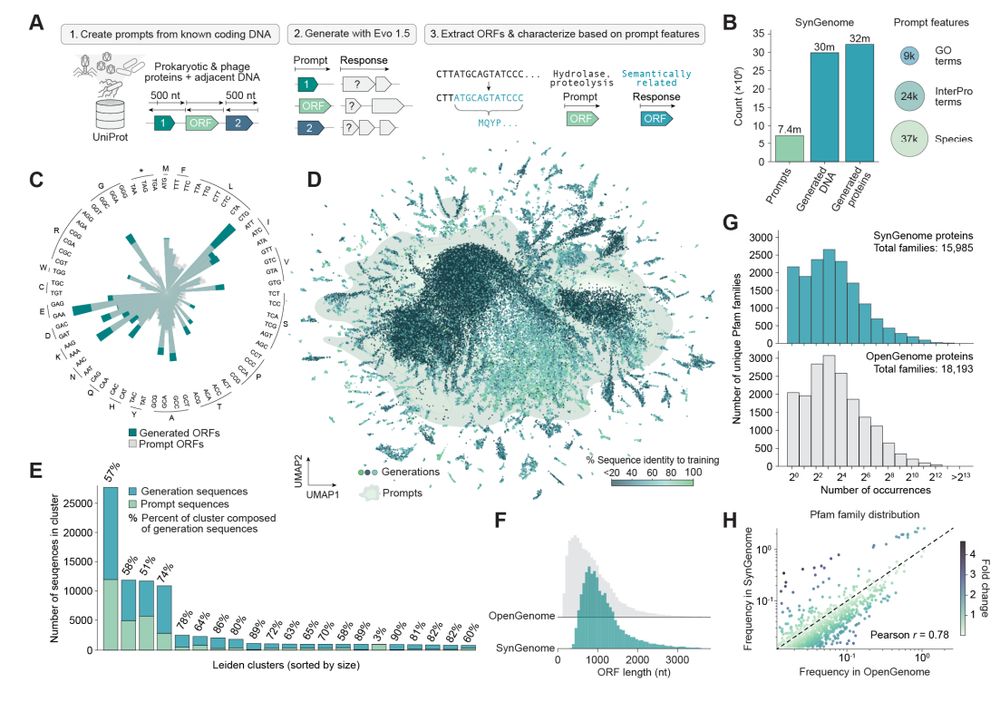
120 billion base pairs of AI-generated genomic sequences with SynGenome.
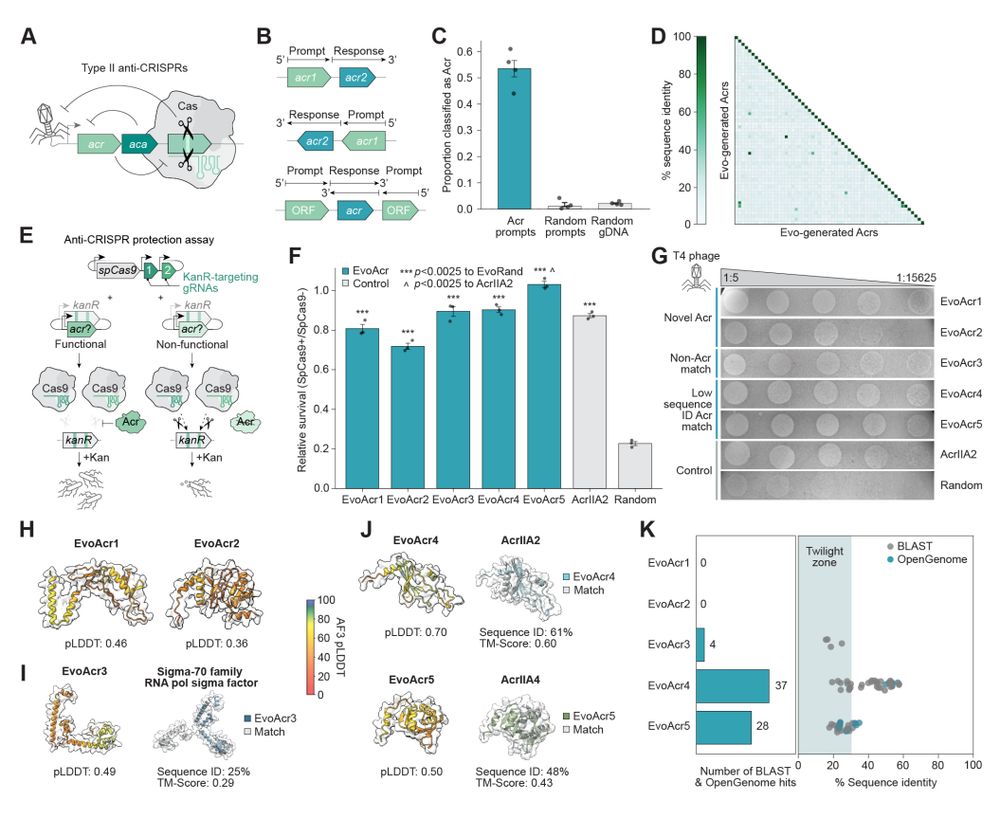
Evo generates functional anti-CRISPR proteins with no homology to known proteins.
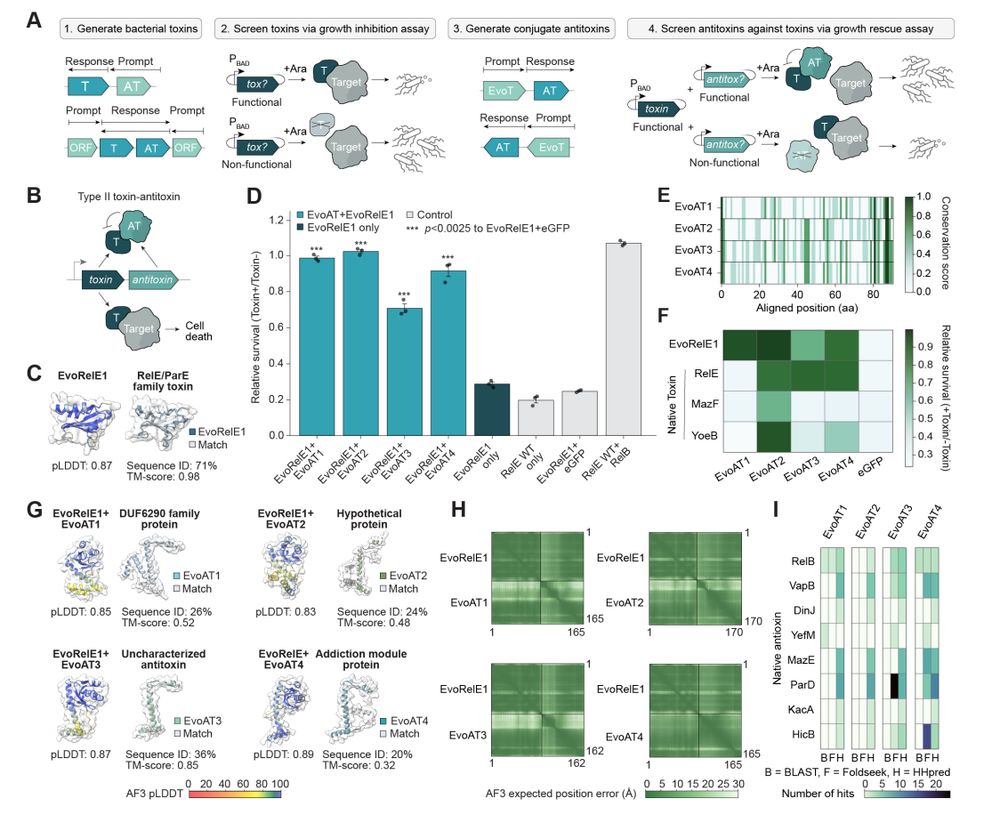
Evo generates functional toxin-antitoxin protein-protein interactions with remote homology to nature.
A protein's position in the genome in relation to other genes is informative about function, so a genomic language model can be prompted to generate
- toxin/antitoxin pairs
- anti-CRISPR proteins
+ a database of 120B synthetic base pairs
www.biorxiv.org/content/10.1...
@brianhie.bsky.social
18.12.2024 22:14 — 👍 60 🔁 19 💬 1 📌 0
🚨 Exciting research alert! 🚨
New #Proteomics study reveals ferroptosis as a key player in Fabry Disease. Using iPSC-derived podocytes, the team identified ALOX15 and ferroptosis as crucial in disease pathology. Could targeting ferroptosis unlock better treatments? 🌟
Details: shorturl.at/G0FHr
11.12.2024 02:23 — 👍 6 🔁 0 💬 0 📌 0
OUTSTANDING PAPER !!!!!
11.12.2024 02:09 — 👍 2 🔁 0 💬 0 📌 0
ProHap: A New Tool for Capturing Genetic Diversity in Proteomics
One proteome to rule them all? We have published a tool to create custom protein databases from large panels of genotypes in @naturemethods.bsky.social. Check the post below to see why it matters, why you should use it, and how it will make your life both easier and more difficult.
10.12.2024 03:41 — 👍 10 🔁 5 💬 2 📌 1

“The genetic architecture of protein stability” 🧪🧶🧬
An additive fitness prediction model trained on ≤2 mutations predicts fitness effects of 3-13 substitutions with R2=0.5
www.biorxiv.org/content/10.1...
28.10.2023 05:49 — 👍 8 🔁 3 💬 0 📌 0
Podcasting help: @proteomicsnews.bsky.social and I record shows remotely using Squadcast, and that works great. But as conferences approach we’ve talked about recording in-person. But I don’t know exactly how. Do we just hook up a bunch of mics and splitters to a MacBook? tascam? mixing board? Help!
06.10.2023 12:06 — 👍 5 🔁 4 💬 1 📌 0
I'm especially excited about this year's courses. The entire curriculum was shaken up, almost every course removed to make room for the new vision and directions of human proteome research! 🧪 🧬
04.10.2023 21:04 — 👍 8 🔁 2 💬 1 📌 0
I discovered this at the skyline course and have been trying to get my workflows into a form that I can use the MS stat's shiny to test the power
05.10.2023 00:03 — 👍 2 🔁 0 💬 0 📌 0
I recently developed a workflow for studying Myc levels and all its related proteins via tmt, worked quite well but I think that work is going to take along time to get published 😀 cool paper btw!
04.10.2023 23:55 — 👍 2 🔁 0 💬 1 📌 0
Thank you! Scored one many thanks to @justinwwalley.bsky.social
04.10.2023 23:52 — 👍 2 🔁 0 💬 1 📌 0
Good blog post about how to approach the analysis of a two species proteomics experiment.
github.com/pwilmart/Hum...
04.10.2023 23:19 — 👍 8 🔁 2 💬 0 📌 0
Damn I didn't realise yall had one!!!! I could have came for once 😅
Also was reading about those enzymes last night very cool
04.10.2023 23:35 — 👍 0 🔁 0 💬 0 📌 0
🧪Analytical chemist | Passionate about mass spectrometry, non-targeted analysis, cheminformatics, and coding | Researching PFAS, natural organic matter, and other complex mixtures | CSIRO Environment | #TeamMassSpec
Mass spectrometry, proteomics, Midwesterner. Views are my own.
Precision-function-based microbiome trials.
Ex vivo microbiome assays.
Microbiome–xenobiotic interactions.
Meta-omics analyses
Empowering Researchers to Focus on Breakthroughs.
Specializing in grant writing support to help researchers secure funding and draft articles. Expertise in chemical R&D, lab automation, impactful scientific communication. (https://www.mbp-rnc.com/)
Dad to four humans and three shih tzus. Professor of Chemistry, mass spectrometrist, physical chemist, associate dean of science.
Bayerisches Zentrum für Biomolekulare Massenspektrometrie | TUM School of Life Sciences | Proteomics | Metabolomics | Bioinformatics | 🇩🇪
Partner site of @diabresearch-dzd.bsky.social, @www.helmholtz-munich.de satellite & @tudresden.bsky.social associate. Basic, pre-clinical & translational beta cell research. Posts by Mark Leaver and Layanne Abu-Bader
Web: https://www.plid.de
Junior Faculty at the Pediatrics and Rare Diseases Group, Sanford Research, Sioux Falls, SD 🦬. #Ubiquitin #RareDiseases #SignalTransduction #StemCells. Chilean 🇨🇱.
https://research.sanfordhealth.org/researchers-and-labs/bustos-lab
Measurements want to be accurate;
Experiments want to be elegant;
Data wants to be beautiful and Data wants to be free
#proteomics
#rstats
The official channel for the British Society for Proteome Research
Post Doctoral Research Fellow | Macquarie University #Glycomics #Glycoproteomics #TeamMassSpec
#Cellzome #TeamMassSpec #Proteomics opinions are my own
Enabling Researchers to Focus on Breakthroughs—
Specializing in proposal writing to secure funding & drafting articles to communicate their work. Expertise: R&D, lab automation, scientific writing (https://www.mbp-rnc.com)
Research pathogenesis of infectious disease & use plasma nanotechnology to improve disease diagnostics & vaccine efficacy
PhD Medicine & Therapeutics (immunology)
Uni Sydney & culturon.com.au
🇬🇧🇦🇺 wife mum reader crafter
Views my own (she/her)
PhD Candidate in the Li Lab at UW-Madison studying neuropeptidomics via mass spectrometry. Advocate for diversity in STEM. #FirstGen (She/her/hers)
AI & Proteomics Researcher @ MannLab - Max Planck Institute | PhD Biochemistry | Bioinformatics | Mass Spectrometry Expert | Automating complex workflows in life sciences
Technical Director CHOP-PENN Proteomics Core Facility, Proteomics, Mass Spectrometry, Analytical Chemistry, Mexican Food addict 🏳️🌈
Systems biologist • Prof at Max Perutz Labs & MedUni Vienna • dad×2 • he/him
#LoveVirology #TeamMassSpec #dataviz
📍 Vienna 🇪🇺
PhD student at UniFr - Switzerland
Exploring the depths of autophagy through mass spectrometry-based proteomics







