the assignment operator is a function and you can overwrite it in the global scope with '<<-' if you need to brick a session:
> unlockBinding('<-', baseenv())
> '<-' <<- function(x) 5
> a = '<-'()
> a
[1] 5
the assignment operator is a function and you can overwrite it in the global scope with '<<-' if you need to brick a session:
> unlockBinding('<-', baseenv())
> '<-' <<- function(x) 5
> a = '<-'()
> a
[1] 5
didn't apply this year so I have no personal point of comparison but always "fun" watching the collapse in real time
10.02.2026 10:30 — 👍 1 🔁 0 💬 0 📌 0wow this really looks like a mountain of LLM generated reviews and/or proposals
10.02.2026 10:29 — 👍 1 🔁 0 💬 1 📌 0(this is a joke but the funding system is rapidly becoming a net negative on the time/money spent vs. funding available axis)
09.02.2026 09:02 — 👍 2 🔁 0 💬 0 📌 0maybe phd programs should skip the research training part and train people to write applications, instead, since that's all you'll have time to do anyway
09.02.2026 09:02 — 👍 3 🔁 0 💬 1 📌 1With a +64% increase in MSCA 2025 scholarship applications, funding cut-offs reached absurd levels. (e.g. a score of 96/100 remained unfunded, got news today from a candidate I sponsored). When rating is this compressed, a lottery among top-rated proposals deserves serious consideration I think.
09.02.2026 07:34 — 👍 14 🔁 5 💬 2 📌 2At long last, my final PhD chapter is out: we developed a novel evolutionary simulator of bacterial pangenomes, Pansim, fitting it to data from >600K genomes using a likelihood-free framework, PopPUNK-mod, to explore neutral and adaptive pangenome dynamics www.biorxiv.org/content/10.6...
07.02.2026 10:08 — 👍 45 🔁 18 💬 2 📌 1
Bioinformatics orphanage - repository of bioinformatics packages that are no longer maintained upstream but still have users, or is a dependency for other more frequently used packages.
I think this is a fun idea, and potentially an important one for the future...💻🧬
github.com/bioinformati...
Very excited about this latest work led by @jermp.bsky.social! Since it's initial release, SSHash has served as the basis for several other tools (Fulgor, piscem, etc.). It was already very fast. It is now *substantially* faster!
www.biorxiv.org/content/10.6...
"Machine learning-based lineage prediction from antimicrobial susceptibility testing phenotypes for Escherichia coli sequence type 131 clade C surveillance across infection types" Spoiler: the MDR ST131-C is pervasive in UTIs, underlying BSI trends. doi.org/10.1099/mgen... @microbiologysociety.org
20.01.2026 09:49 — 👍 4 🔁 6 💬 1 📌 0
Fluorescence increases over time in mother cells due to aggregates.
How does E. coli age? According to this #mBio study, the the decisive factor driving growth decline in E. coli is not the presence of protein aggregates, but the fraction of the intracellular space they occupy. Learn more: asm.social/2KZ
07.01.2026 17:38 — 👍 12 🔁 11 💬 1 📌 0
📣 preprint alert!
We sampled #klebsiella isolates from💩 patients when they entered the hospital & 🩸 after development of infection.
🎯 We tracked phenotypic changes & correlate them w genotypic changes.
@klebclub.bsky.social
-in collab with Clermont-Ferrand lab
#microsky
🧵
doi.org/10.64898/202...

The changing roles of Escherichia coli www.nature.com/articles/s41... #jcampubs
24.12.2025 21:23 — 👍 7 🔁 5 💬 0 📌 0
"..based on a common wavefront design that can be adapted to support a variety of dynamic programming algorithms: local, global, and semi-global alignment of genomic and protein sequences with a variety of commonly used scoring schemes" from
@martinsteinegger.bsky.social andco

"Ethical standards do not become optional because the participants are African."
bktitanji.substack.com/p/how-unethi...

I wrote a piece on LLMs, citation metrics, and the institution of science. It's been up for a couple of days, but it is officially "out" now.
15.12.2025 16:19 — 👍 176 🔁 61 💬 7 📌 14
Preprint Alert!
With @tmthrz.bsky.social and @rayanchikhi.bsky.social we aim to tackle practical unitigs compression!
A thread:
If you ever need to fuzzy search some DNA, sassy is your tool.
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
Mapping the evolutionary path towards multi-drug resistance in the pandemic Escherichia coli ST131 lineage https://www.biorxiv.org/content/10.64898/2025.12.08.692488v1
10.12.2025 14:31 — 👍 2 🔁 2 💬 0 📌 0
Lovely to feature alongside the brilliant Birgit Sattler & Tom Battin in this story about life in Earth's dying ice.
www.the-scientist.com/the-ice-is-a...
Amazing dataset describing the metabolic profiles of 66 E. coli transporter knockouts by @sulheim.bsky.social with Peter Doubleday and @nzamboni.bsky.social. The tool, set up by @lambdapp.bsky.social and Eric Ulrich allows you to explore for yourself. Enjoy! Pre-print here: doi.org/10.1101/2025...
03.12.2025 13:11 — 👍 27 🔁 17 💬 0 📌 0
Colibactin-DNA interstrand crosslinks structure reveals DNA-damaging acitivity of colibactin that is linked to colorectal cancer #MicroSky www.science.org/doi/10.1126/...
04.12.2025 22:07 — 👍 21 🔁 10 💬 1 📌 1
1/9 Just out:
k-mer indexes are the backbone of fast search in genomic data, but many degrade under small k, subsampling, or high diversity.
With Ondřej Sladký and @pavelvesely.bsky.social we asked: can we build one that works efficiently for any k-mer set?

Preprint out! Check out our new long-read metagenomic SNP-caller, SNooPy 😀. Work with Chris Quince. Thread 🧵
👉 www.biorxiv.org/content/10.6...


🧬🛡️How are new immune mechanisms created?
We show how Lamassu antiphage system, originated from a DNA-repair complex and evolved into a compact and modular immune machine, wt Dinshaw Patel lab in @pnas.org.
👏 @matthieu-haudiquet.bsky.social, Arpita Chakravarti & all authors!
doi.org/10.1073/pnas...
Optimized k-mer search across millions of bacterial genomes on laptops https://www.biorxiv.org/content/10.1101/2025.11.23.690050v1
26.11.2025 16:47 — 👍 26 🔁 13 💬 0 📌 1
If you're about to push these last manuscripts before wrapping up 2025, remider that our MGen Eukaryotic collection @microbiologysociety.org is open & growing; #Protists #Parasites #Fungi; friend or foe; all welcome 😊
#MicroSky #IDSky #Protistsonsky 🧬💻
www.microbiologyresearch.org/content/micr...
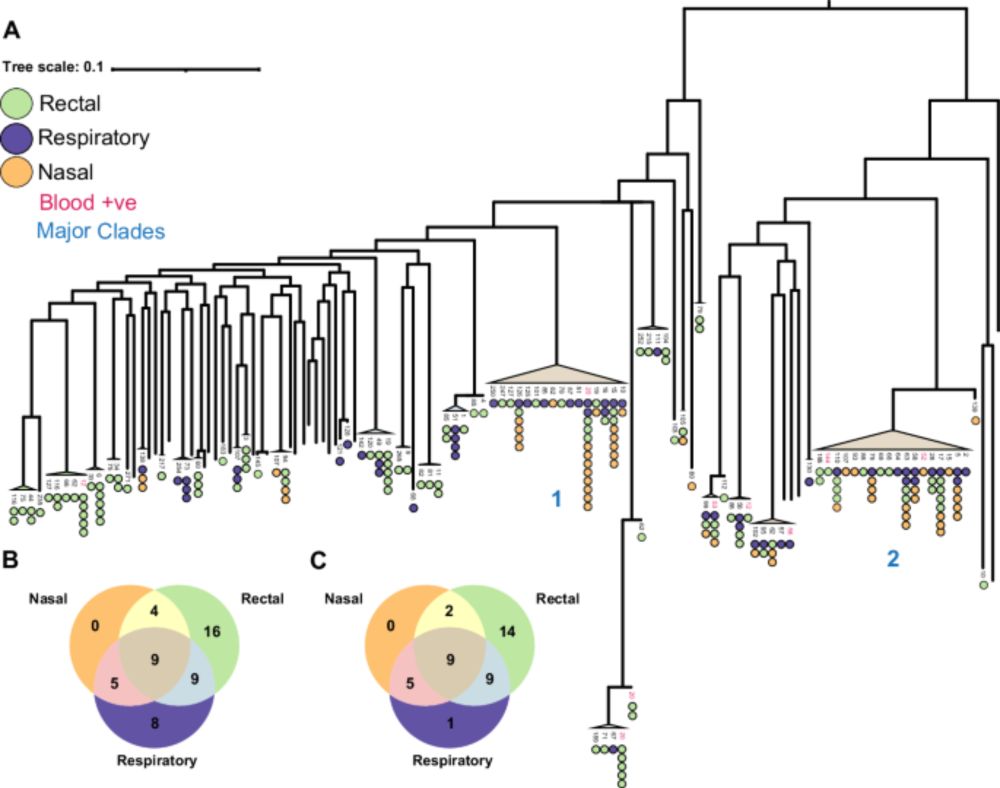
Really pleased to share the first paper to come out of the lab.
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...

#microsky #phagesky #phage
Anyone who’s tried deleting prophages in the lab by HR knows the difficulties of the task. Here we have an example of how HR-mediated natural transformation might hit the same hurdle in a more native context.
academic.oup.com/mbe/article/...

Hot off the press! Our latest paper led by @fernpizza.bsky.social, understanding how plasmids evolve inside cells. These small, self-replicating DNA circles live inside bacteria and carry antibiotic resistance genes, but also compete with one another to replicate. 1/
www.science.org/doi/10.1126/...