🚨 EntoSieve is now published in Molecular Ecology Resources: Fast and accurate size-sorting of bulk insect samples! Perfect for boosting DNA megabarcoding & metabarcoding projects. 🧬🔬 DIY low-cost device, gentle on specimens and yet efficient 🦋🐞 here: onlinelibrary.wiley.com/doi/full/10....
14.03.2025 15:03 — 👍 31 🔁 9 💬 2 📌 2
No, just regular 2 mL screwcap tubes.
31.01.2025 10:13 — 👍 1 🔁 0 💬 0 📌 0

GitHub - DominikBuchner/apscale: Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data
Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data - DominikBuchner/apscale
Just released version 1.6.0 of the APSCALE pipeline. This update includes improved log writing and added failsafe features for better handling of invalid settings. Check it out on GitHub:#apscale #metabarcoding #pipeline
github.com/DominikBuchner…
06.01.2023 15:23 — 👍 4 🔁 3 💬 0 📌 0
RNA cleanup with magnetic beads
This protocol describes cleaning up RNA extracts with carboxylated magnetic beads and a PEG-NaCl buffer. It can also be used for volume reduction of a sample or buffer exchange...
We're happy to share our RNA cleanup protocol on @protocolsIO! Our method is significantly more cost-effective than commercial kits, without sacrificing effectiveness. Check it out here:#RNA #cleanup #openscience @leeselab
protocols.io/view/rna-clean…
05.01.2023 06:46 — 👍 0 🔁 0 💬 0 📌 0
I hope my protocol will be a useful resource for the science community. I'm always looking for ways to give back, and I just wanted to say happy holidays to all of you. Merry Christmas! #merrychristmas #sciencecommunity #protocolsio
23.12.2022 09:20 — 👍 0 🔁 0 💬 0 📌 0
Hint: Cleaning a phenol-contaminated centrifuge is not exactly a fun adventure. Avoid the hassle by taking good care when working with phenol-chloroform. Trust me, your labmates will thank you. #labtips #protocolsio 5/5
23.12.2022 09:15 — 👍 0 🔁 0 💬 1 📌 0
Our protocol is a reverse-engineered version of the RNeasy PowerSoil Total RNA Kit, but with a fraction of the cost. Our method reduces the cost from ~27€ per prep to ~6€, making it more accessible for researchers on a budget. #soilmicrobiology #RNA #costsaving #protocolsio 2/5
23.12.2022 09:15 — 👍 0 🔁 0 💬 1 📌 0
 12.12.2022 16:05 — 👍 0 🔁 0 💬 0 📌 0
12.12.2022 16:05 — 👍 0 🔁 0 💬 0 📌 0
Twitter, your help is needed:
I'm desperately searching for small-volume, gravity-flow anion exchange columns comparable to the JetStar columns in the Qiagen RNeasy PowerSoil Total RNA Kit. Does anyone know a supplier for those columns?
30.11.2022 05:20 — 👍 0 🔁 0 💬 0 📌 0
Soon the protocol will be added to my @protocolsIO. Still really hate working with phenol-chloroform but the result is worth the struggle.
18.11.2022 16:23 — 👍 0 🔁 0 💬 0 📌 0
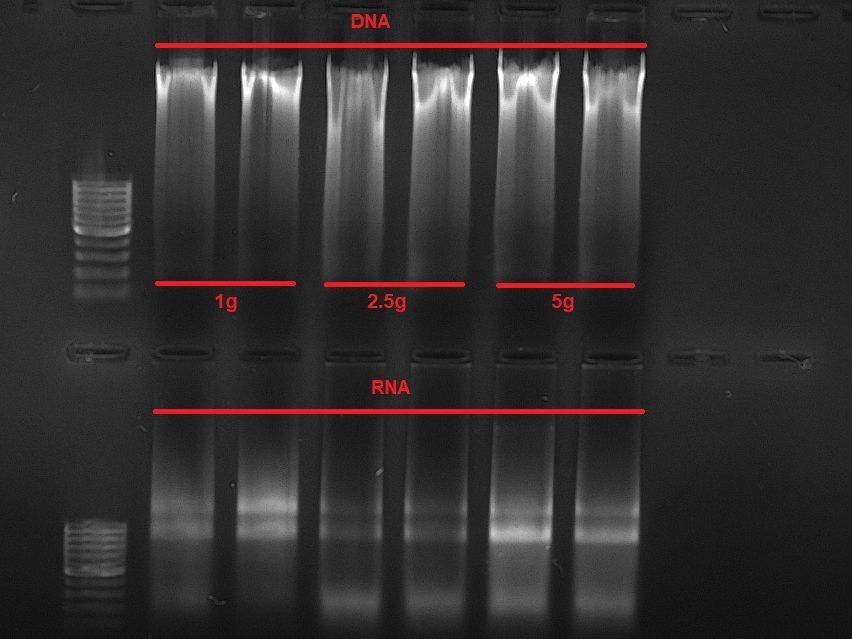
Co-extracting DNA/RNA from sediment haunted me for almost 2 years now with limited success so far. This changed today: I finally developed a protocol to process up to 15g of sediment in one extraction while splitting up DNA/RNA along the way. Will sleep like a baby tonight.
18.11.2022 16:21 — 👍 0 🔁 0 💬 2 📌 0

It's working 😍
10.11.2022 16:45 — 👍 0 🔁 0 💬 0 📌 0

Sales representative:"How many plates do you need?"
Me:"Yes"
#upscaling
24.10.2022 14:02 — 👍 0 🔁 0 💬 0 📌 0
Dominik Buchner on protocols.io
Metabarcoding, Genetics, Bioinformatics, Robotics
Since everyone is talking about open science these days and I fully agree that knowledge should be accessible to anyone I will start putting all protocols I know on @protocolsIO. DNA extraction and bead cleanup are already up. Will add more gradually. 👨🔬🙂
protocols.io/researchers/m4…
13.10.2022 14:04 — 👍 0 🔁 0 💬 0 📌 0
Update on #BOLDigger: For the selection of any type of top hit all database entries containing punctuation and/or numbers will be ignored. This leads to cleaner taxa lists and less data cleaning needed for final analysis 🤓
16.09.2022 11:14 — 👍 0 🔁 0 💬 0 📌 0
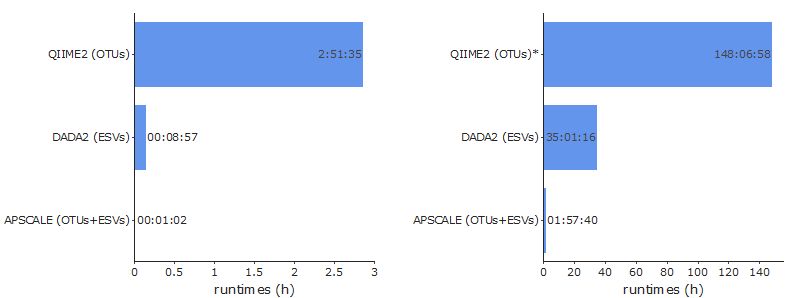
While producing similar results we were able to boost performance almost tenfold compared to other well known pipelines. APSCALE scales well with dataset size in terms of runtimes and ressources needed. Make sure to check out the benchmark!
29.08.2022 05:36 — 👍 0 🔁 0 💬 0 📌 0

When the mainboard of your (real) computer is determined to commit suicide, so you are forced to spend the last weekend of your vacation backing up data and disassembling it. Good excuse to buy a new computer though 🥲🥲 #goodbyeoldfriend
07.08.2022 17:09 — 👍 0 🔁 0 💬 0 📌 0
Updated BOLDigger and BOLDigger command-line to version 2.0.0. Selection of the top hit is now more sophisticated, check out the Readme for a detailed description of the function. Command-line version is now as quick and functional as the GUI. #BOLDigger #python #metabarcoding
24.05.2022 16:57 — 👍 0 🔁 0 💬 0 📌 0

Started to work on automatic image annotation as a side project. Got serious quickly. Ordered an LED table and took photos with my cell phone. Works pretty well already, still need to add a scale to measure insects in mm instead of pixels 🤓#python #opencv #malaise @leeselab
16.05.2022 15:32 — 👍 0 🔁 0 💬 0 📌 0

Time to learn some new (and hopefully exciting) stuff. I promise to stop using this as soon as I have my PhD. Good to know that this will still take a while 😁 #coding #ai #python
09.05.2022 16:01 — 👍 0 🔁 0 💬 0 📌 0
#BOLDigger update: Asynchronous requests are now implemented into #BOLDigger speeding up the identification engine 2-4 fold, depending on internet connection and database availability. Up to 1000 sequences per hour are now possible! @leeselab
14.02.2022 13:26 — 👍 0 🔁 0 💬 1 📌 0

GitHub - TillMacher/apscale_gui: Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data
Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data - TillMacher/apscale_gui
Available as a command-line version as well a GUI-version integrated by @TillMacher. Feel free to try it out and give us feedback on what options you would like to be added. #metabarcoding #pipeline #Bioinformatics
github.com/TillMacher/aps…
18.01.2022 17:32 — 👍 0 🔁 0 💬 0 📌 0
GitHub - DominikBuchner/apscale: Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data
Advanced Pipeline for Simple yet Comprehensive AnaLysEs of DNA metabarcoding data - DominikBuchner/apscale
Check out our metabarcoding pipeline #apscale. Finally found the time to close the gap between my demultiplexing tool and #BOLDigger. Easy to install, option to run an all-in-one analysis from demultiplexed read to the OTU/ESV table. @leeselab
github.com/DominikBuchner…
18.01.2022 17:32 — 👍 0 🔁 0 💬 1 📌 0
All I want for Christmas is ~ 500 GB of raw read data or ~ 3.5 billion PE reads to demultiplex. #merrychristmas @leeselab #malaisetraps #bigdata
24.12.2021 16:27 — 👍 0 🔁 0 💬 0 📌 0
Molecular ecologist working on #Multiomics and #Microbiome research. SUCCeSS Postdoc fellow @AcesSthlmUni. Views my own.
linktr.ee/JoeselleSerrana
Research tech at the Centre for Integrative Biodiversity Discovery @mfnberlin.bsky.social 🪰🦟🐝🧬🧪
Tell your cat I said pspspsps
He/him 🏳️🌈
Biodiversity science in the 21st century: Diptera, Natural History, Phylogenetics, Oxford Nanopore, genomics, metagenomics, NGS barcoding, metabarcoding.
Professor at University of Duisburg-Essen 🇪🇺🇩🇪 tweeting mostly about the work we do in our Aquatic Ecosystem Research Group. A lot of DNA / eDNA & popgen stuff on streams & automated biodiversity monitoring approaches.
🔬 PhD student @leeselab
| Molecular ecologist working on #biodiversity monitoring using #eDNA #metabarcoding | 📈 @trend_dna
project 🧬
Nature photographer 📷
Postdoctoral researcher @ University of Duisburg-Essen
| #eDNA #parasites #freshwater @impact-parasites.bsky.social
| Studying biodiversity & ecology with #DNA
Researcher @Naturalis.bsky.social, Assistant Professor
@UniLeiden. @leeselab.bsky.social alumnus. Utilizing molecular tools to study communities and holobionts.
Co-Founder, SparkPR | AI, Tech, BioTech, VC, Longevity | Citizen Scientist & Advocate | Mom to twin angels Addi & Cassi with Niemann Pick Type C (2004-2019)
Full Professor of Aquatic Ecology University Zurich & @eawag.bsky.social | Kuratorium Forum Biodiversity | Founding Director https://biodiversitaet.uzh.ch/en.html | all & beyond #biodiversity, #eDNA & #AquaticEcology | Like=interesting
www.altermattlab.ch
Lab Rat Extraordinaire, Plant Virologist, Human. Part of the http://cell-free.com and http://liberumbio.com team!
Fascinated by freshwater biodiversity, combining morphology & #eDNA-based methods for #monitoring & #assessment 🔬🧬| 🇲🇽 in 🇱🇺🇪🇺via 🇨🇷🇩🇪🇸🇪 | 🏳️🌈
• PhDc at Jackson’s Aquatic & Knowles’s Microbiome Ecology Labs @OxfordBiology.bsky.social
• #firstgen Latina in STEM
• University of Buenos Aires alumna 🇦🇷
• My views
Fascinated and humbled by #biodiversity. I understand it using #eDNA as CEO at SimplexDNA. Views and typos are my own. She/her(s).
Postdoctoral researcher at Linnaeus University Sweden | New Zealander | Ecologist
Head of Virology at Charite - Universitätsmedizin Berlin
Professor at The University of Hong Kong (www.seymourlab.net). I am an eDNA specialist with a background in ecology, biogeography and metacommunity dynamics. My lab focuses on marine and freshwater systems. Associate editor for Environmental DNA
Postdoctoral Fellow in University of Victoria
Environmental DNA | Environmental monitoring | Community ecology
https://linktr.ee/marklouiedlopez
Associate Professor in Aquatic Biology @tcddublin Trinity College, The University of Dublin. FTCD.
The 3rd most popular 3rd of View From The Ninian





 12.12.2022 16:05 — 👍 0 🔁 0 💬 0 📌 0
12.12.2022 16:05 — 👍 0 🔁 0 💬 0 📌 0









