Our lab is seeking candidates to apply for a 3-year postdoctoral fellowship in the area of plant synthetic biology funded by the Spanish Research Council. Deadline for applications is 17 Dec. Please, spread the word! contact: k.wabnik@upm.es
18.11.2025 11:11 — 👍 3 🔁 3 💬 0 📌 0
Wow cool! COngratz Ari!
30.10.2025 21:14 — 👍 0 🔁 0 💬 1 📌 0

Fig. 4.Model of auxin de-repression. When the auxin concentration in the nucleus is low (A), the effect of ARFas, as well as other non-ARF transcription factors, is limited by Aux/IAA-mediated recruitment of the TPL co-repressor, closing the locus. As auxin levels rise (B), AFBs ubiquitinate Aux/IAAs, thereby allowing all transcription factors to act on the locus. As a result, there is a significant difference in gene expression between baseline and auxin induction. In afb1,2,3,4 mutants (C), Aux/IAAs are not degraded, and therefore the promoter remains closed. In the iaa2mDII degron mutant (D), one of three Aux/IAAs is stabilized, thereby resulting in only partial promoter closure. In arfasept lines, Aux/IAAs are not recruited to the promoter. While gene expression remains diminished without activating ARFs, other non-ARF transcription factors are allowed to provide baseline gene expression. However, there is no auxin-mediated induction.
🌱📖 RESEARCH 🌱📖
Bascom et al. used CRIPSR/Cas9 to produce a septuple AUXIN RESPONSE FACTOR mutant in the model bryophyte Physcomitrium patens , revealing the developmental consequences of the complete loss of auxin-mediated gene activation 🌱📖
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
29.09.2025 10:42 — 👍 20 🔁 7 💬 1 📌 1

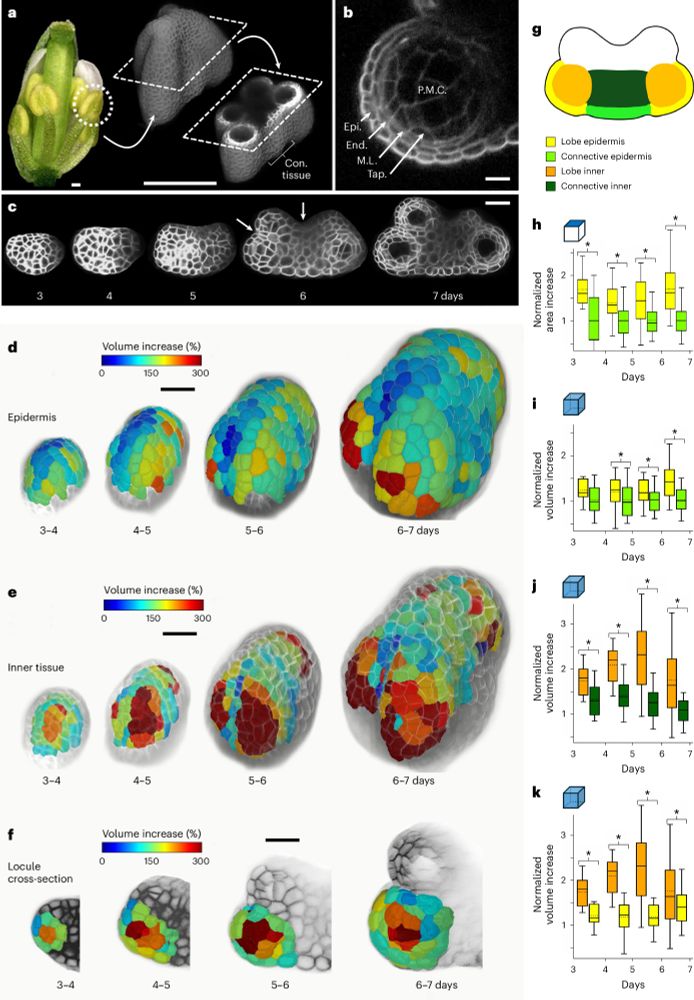
Fig. 1 (shortened, full legend in paper): Auxin responses and auxin-adjacent transcription factors (TFs) control developmental transitions in bryophyte gametophytes. (A) Wild-type (WT) auxin responses (left) cycling between low and high auxin. Under low auxin, A-ARFs are kept in a repressive complex by AUX/IAAs via recruitment of the TOPLESS co-repressor and associated histone deacetylases (HDACs). Under high auxin, auxin mediates the interaction between TIR1/AFB and AUX/IAAs, leading to AUX/IAA ubiquitination and proteolysis. A-ARFs are then free to interact with SWI/SNF ATPases and histone acetylases (HACs), leading to open chromatin states. Positive (TIR1/AFB, A-ARF, SWI/SNF, HAC), negative (AUX/IAA, TPL, HDAC) and auxin-adjacent (TFs) factors are shown. Auxin is represented by an A.
🌱🧬 INSIGHT 🧬🌱
Complete loss of A-ARF activity in bryophytes reveals a role for auxin-adjacent regulatory networks – Flores-Sandoval & Bowman comment on an original research article by Bascom et al.
📝 Insight: doi.org/10.1093/jxb/...
🔬 Research: doi.org/10.1093/jxb/...
#PlantScience 🧪
22.09.2025 13:32 — 👍 9 🔁 6 💬 0 📌 0

🌱 #SeminarSeriesCBGP is back! 🎙️
Join us Sept 2025–Jan 2026 for top voices in plant science at CBGP
📍 CBGP Auditorium + Zoom
🕛 12:30 PM
🔗 Speakers & calendar: shorturl.at/Atsyn
📎 Full schedule w/ Zoom links: shorturl.at/yT3e6
Don’t miss it! 👩🔬👨🔬
19.09.2025 08:50 — 👍 5 🔁 2 💬 0 📌 1

#PaperCBGP
🌱🔬 Plant cell walls are more than physical barriers. They are dynamic shields!
A CBGP review published in @jxbotany.bsky.social explores how cell walls shape plant defense against pests, paving the way for more sustainable farming strategies ♻️🌍
📎 More: shorturl.at/GLNYL
10.09.2025 11:39 — 👍 9 🔁 5 💬 0 📌 0
#ICAR2025
17.06.2025 17:27 — 👍 0 🔁 0 💬 0 📌 0
Check out our lab posters: P125 and P88 #ICAR25 #PlantScience @cbgpmadrid.bsky.social To learn about New 4D modeling simulators and auxin-dependent root growth!!
17.06.2025 17:26 — 👍 2 🔁 1 💬 1 📌 0

Join the PlantDynamics to develop next-gen 4D organ simulators
The @PlantDynamics Laboratory (https://pdlab.es) is seeking a motivated and talented
PlantDynamics is recruiting a programmer to join efforts in developing plant (and other) organ simulators
euraxess.ec.europa.eu/jobs/341946
Contact me directly if you are interested: k.wabniK@upm.es
08.05.2025 15:11 — 👍 3 🔁 2 💬 0 📌 0

The Camelot paper is now published. If you ever wanted to try biomechanics, Camelot is an inexpensive and easy way to get started. @matmajda.bsky.social
t.co/x3l0lllNS2
28.04.2025 08:09 — 👍 13 🔁 11 💬 0 📌 0


We have a new Doctor form the #pollmannlab in town 👨🏻🎓Congratulations to Adrián to his wonderful PhD defense 🎉 Outstanding job. Also a great shoutout to the panel for the critical assessment of the work. We will surely pick up some of the points.
@cbgpmadrid.bsky.social @etsiaabupm.bsky.social @upm.es
11.04.2025 16:36 — 👍 7 🔁 3 💬 1 📌 0

Our editors write:
- Cell polarity: Unequal signalling and biosynthesis in daughter cells rdcu.be/egIl5
07.04.2025 11:50 — 👍 17 🔁 5 💬 0 📌 0
de nada!
30.03.2025 19:43 — 👍 1 🔁 0 💬 0 📌 0
Research Briefing by @routierlab.bsky.social and @sysilveira.bsky.social about our recent paper @natureplants.bsky.social
28.03.2025 18:48 — 👍 5 🔁 1 💬 0 📌 0
Fantastic collaboration! and exciting findings! congratz to all involved!
10.03.2025 22:19 — 👍 2 🔁 0 💬 0 📌 0

🌱💡 Computational modeling and genetic experiments confirm that this asymmetry optimizes root growth, ensuring robust meristem function. @wabniklab.bsky.social
10.03.2025 14:58 — 👍 4 🔁 1 💬 2 📌 0

Time-lapse imaging back then, Zeiss scope from 1950. #microscopy #timelapse #imaging
28.02.2025 17:36 — 👍 2 🔁 0 💬 0 📌 0
Thx!
21.02.2025 09:36 — 👍 1 🔁 0 💬 0 📌 0
Using computer modeling and quantitative analysis of plant transcriptomes, we show how common design principles in core clock control photoperiod-dependent florigen function in short-day vs long-day plants #clock #circadianclock #oscillators #flowering #plantgrowth
21.02.2025 06:23 — 👍 2 🔁 0 💬 0 📌 0
very neat! Congratulations!
20.02.2025 22:10 — 👍 0 🔁 0 💬 0 📌 0
and @ataly.bsky.social @bic-bordeaux.bsky.social
First authors Jessica Perez-Sancho, Marija Smokvarska , Gweenogan Dubois .
20.02.2025 15:59 — 👍 10 🔁 1 💬 2 📌 0
Our lab is looking for a enthusiastic researcher with the master degree (Computational Biology, Computer Science, Game development) to develop next-gen computer games to model organ development in 4D. If you are interested drop me an email: k.wabnik@upm.es
03.02.2025 08:39 — 👍 5 🔁 8 💬 0 📌 0

#SeminarCBGP | 07/02/25 - 12:30h
🧑🏽🔬 Dr. Robertas Ursache
🌍 Centro de Investigación en Agrigenómica (CRAG)
Barcelona, Spain
📝 "The suberinterestingrole of GELP proteins”
💻 Zoom ID: 867 7606 8944 | Código acceso: 189185
#somosUPM
03.02.2025 08:08 — 👍 1 🔁 1 💬 0 📌 1
Our lab investigates how plants respond to abiotic stress and changing environments using auxin and regulated proteolysis.
https://sites.google.com/view/salehinlab-ncat-edu/home
Postdoctoral researcher at Pollmann's lab (CBGP, Madrid)
Plant-microbe interactions 🌱🍄🦠
32. Biochemist. PhD in Plant Biotechnology. Postdoc at CBGP, Madrid. Previous MPIMP, Potsdam. 🍓🍅🌱👨🏻🔬
Faculty @ Mizzou & Danforth Center. Loves brace roots & biomechanics.
A fine group of plant biologists interested in morphogenesis and trying to get to the roots of things. Home base: Heidelberg University - https://www.cos.uni-heidelberg.de/en/research-groups/cell-and-developmental-biology.
Biophysicist, Group Leader at MPIPZ. Studying the multicellular dynamics in plants, combining theory and experiments. Also, fascinated by music and dance.
Canada Research Chair in Biomechanics of Plant Development. McGill University, Montreal, Canada. www.plantbiomechanics.net.
For my administrative role follow me on X at @AnjaGeitmann
Group leader at Wageningen University & Research, NL | Prokaryotic immune systems | Views are my own
Plant cell/developmental biologist @RDP Lyon, loves cell edges, mechanics, and lateral roots
Más de 25 años dedicados a la I+D+i y a la divulgación científica en el campo de la Biología y la Biotecnología de Plantas.
Universitat Politècnica de València - Consejo Superior de Investigaciones Científicas @csic.es
Nature Plants is an online-only, monthly journal publishing the best research on plants (hopefully).
Plant developmental biologist; Researcher at IBPM-CNR, Rome, IT
Cuenta oficial del Centro de Biología Molecular Severo Ochoa (CSIC-UAM)
Plant development & cell biology researcher, stem cells, cell division, cell wall, water response...
Group leader at CAS Center for Excellence in Molecular Plant Science(CEMPS) and CAS-JIC Centre of Excellence for Plant and Microbial Science (CEPAMS)
🌿 Centro mixto de I+D+i | UPM-INIA/CSIC
📌 Parque Científico y Tecnológico de la U.P.M. | Campus de Montegancedo
🔗 https://www.cbgp.upm.es/
Weigel Lab aka WeigelWorld Repost≠endorsement
#OpenScience
COIs http://tinyurl.com/DWCOI2022 GoogleScholar http://tinyurl.com/DeWeScholar
She/ Her, Plant development & Plant Cell Biology. Roots lover. Runner. Professor @uni-freiburg.de @biologyunifreiburg.bsky.social
Plant developmental biologist with a key interest in vascular tissues and cell division orientation at VIB/UGent 🇧🇪🇪🇺🌱