
Submit them to Aurorasaurus!!! They're an NSF funded project out of University of New Mexico that takes crowdsourced aurora observations for research purposes:
www.aurorasaurus.org
@maikemorrison.bsky.social
Building mathematical tools to answer biological questions Ecology & evolution, sometimes microbiomes & cancer Omidyar Fellow @SFIscience.bsky.social Former PhD student with Noah Rosenberg, Stanford University MaikeMorrison.com

Submit them to Aurorasaurus!!! They're an NSF funded project out of University of New Mexico that takes crowdsourced aurora observations for research purposes:
www.aurorasaurus.org



Northern lights over Santa Fe!
12.11.2025 05:27 — 👍 7 🔁 0 💬 0 📌 0
My little brother made the coolest thing you'll see all day www.youtube.com/watch?v=ULpN...
10.11.2025 04:09 — 👍 5 🔁 3 💬 0 📌 0
Glass beads of all sorts balancing on wavy threads.
How is functional variation at large-effect loci maintained in natural populations, even as environments change? In a paper led by @mkarag.bsky.social, we tracked known pesticide resistant alleles in outdoor 𝘋. 𝘮𝘦𝘭𝘢𝘯𝘰𝘨𝘢𝘴𝘵𝘦𝘳 cages & inferred selection and dominance from temporal sequencing data.
06.11.2025 21:51 — 👍 35 🔁 17 💬 1 📌 0
"I voted! / !Yo voté!" sticker featuring a road runner, a yucca, and a Zia symbol in the background
Rank-choice voting AND a roadrunner sticker? NM has voting figured out.
04.11.2025 17:44 — 👍 20 🔁 5 💬 1 📌 0
1/9 Excited to share the preprint for the second half of my PhD with @mollyschumer.bsky.social: Connecting human environmental impacts to hybridization in swordtail fish through genomics, GIS, water chemistry, and histology. Thread below!
#SciSky
www.biorxiv.org/content/10.1...
Due to maintenance, Research.gov (including access to NSF-PAR, GRFP, PES, and ETAP) will be unavailable from Fri., 4/25 at 10:00 PM ET to Sat., 4/26 at 1:00 PM ET. NSF apologizes for any inconvenience.
🚨 Practical URGENT tip for NSF grantees:
Out of an abundance of caution, I would right now go into Research.gov and…
1. Download your NSF award letters.
2. Print PDF your annual reports.
3. Screenshot the status table for annual reports.
NSF is planning maintenance tomorrow to Research.gov

ATTENTION: NSF GRANT RECIPIENTS
We received a heads up from a trusted source that you should proactively download/print/screen shot any documentation on research.gov pertaining to your NSF awards, both those that are current and any that have closed in the last 5-6 years.
1/n

"The silence from the leaders of our university—at least in public—suggests the assault has already produced the intended chilling effect, but together we can overcome it." Good read from Stanford History profs Jessica Riskin and Priya Satia.
stanforddaily.com/2025/03/17/i...

Thanks! Totally! We have a preprint using a similar framework to look at mutational signature heterogeneity across tumors (collab with @nalcala.bsky.social doi.org/10.1101/2023...) and we also used this framework to look at heterogeneity of immune cell infiltration (analysis not published yet).
17.03.2025 18:59 — 👍 0 🔁 0 💬 0 📌 0#mevosky #microsky
14.03.2025 20:51 — 👍 2 🔁 0 💬 0 📌 08/8 FAVA is the culmination of many conversations and collaborations. Thanks especially to my co-authors, @ksxue.bsky.social and Noah Rosenberg–I truly feel like I won the collaborator/mentor lottery. And thanks to you for reading! Paper is here if you want to read more: doi.org/10.1073/pnas...
14.03.2025 20:46 — 👍 7 🔁 0 💬 3 📌 0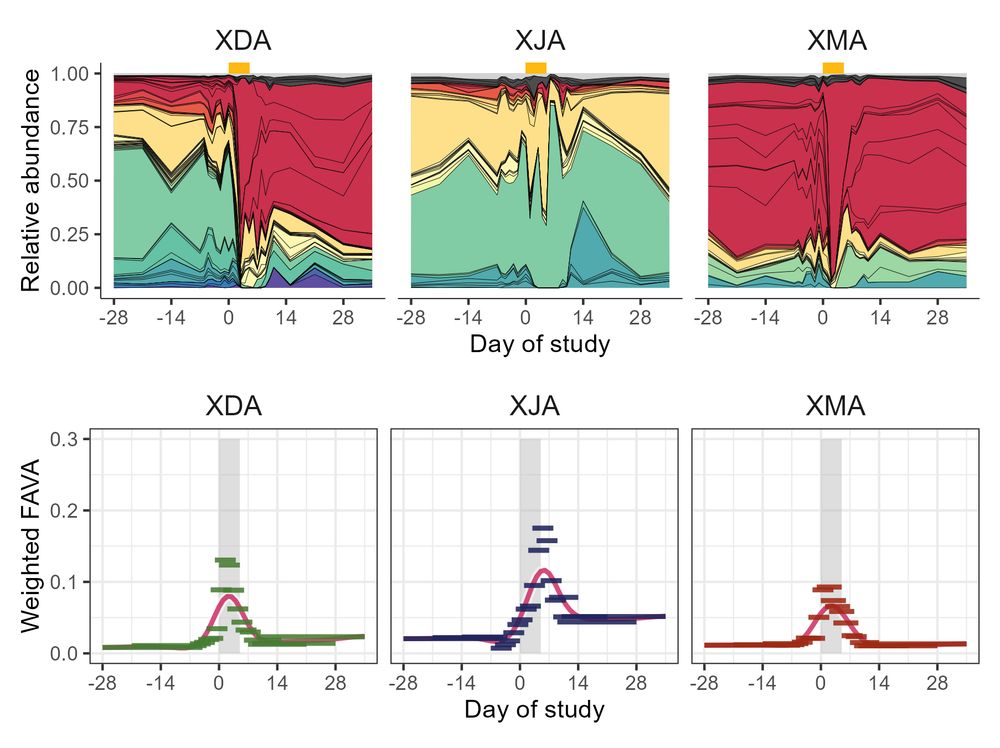
Plot showing longitudinal microbiome composition for three individuals who took an antibiotic, along with corresponding plots showing FAVA computed in sliding windows. For all subjects, FAVA increases during the antibiotic window. In one subject (XDA) we see that FAVA can return to stability (low FAVA) without returning to the pre-abx composition.
7/ (Ex. 2) In longitudinal microbiome samples from healthy humans taking an antibiotic (collab with @ksxue.bsky.social), FAVA is much higher post-abx than pre-abx, and just half of the 22 subjects return to their baseline FAVA level (computed in 6-sample sliding window) during the 30 days post-abx!
14.03.2025 20:46 — 👍 5 🔁 0 💬 1 📌 0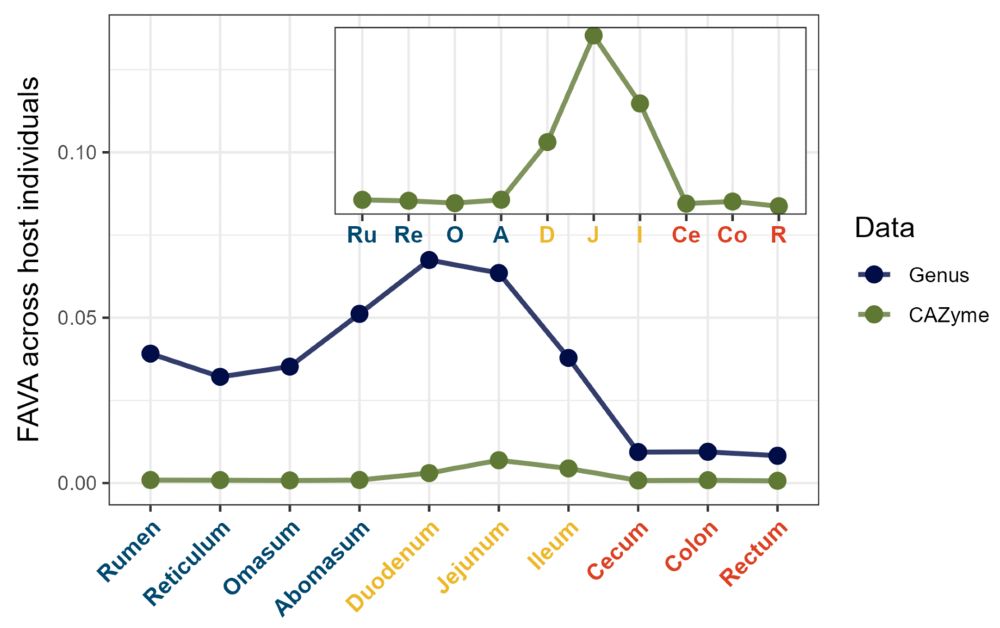
Plot showing FAVA computed with genus or CAZYme abundances across ruminant host individuals. FAVA is much higher for genus than CAZyme abundances. For both data types, FAVA peaks in the small intestine and is quite low in the large intestine, suggesting much variation across individuals would be missed with stool sampling.
6/ (Ex. 1) In microbiome samples collected along the GI tracts of individuals from 7 ruminant species, FAVA peaks in the small intestine–so lots of variability would have been missed by stool sampling! We also find taxonomic variability >> functional variability–evidence for functional redundancy.
14.03.2025 20:46 — 👍 5 🔁 0 💬 1 📌 0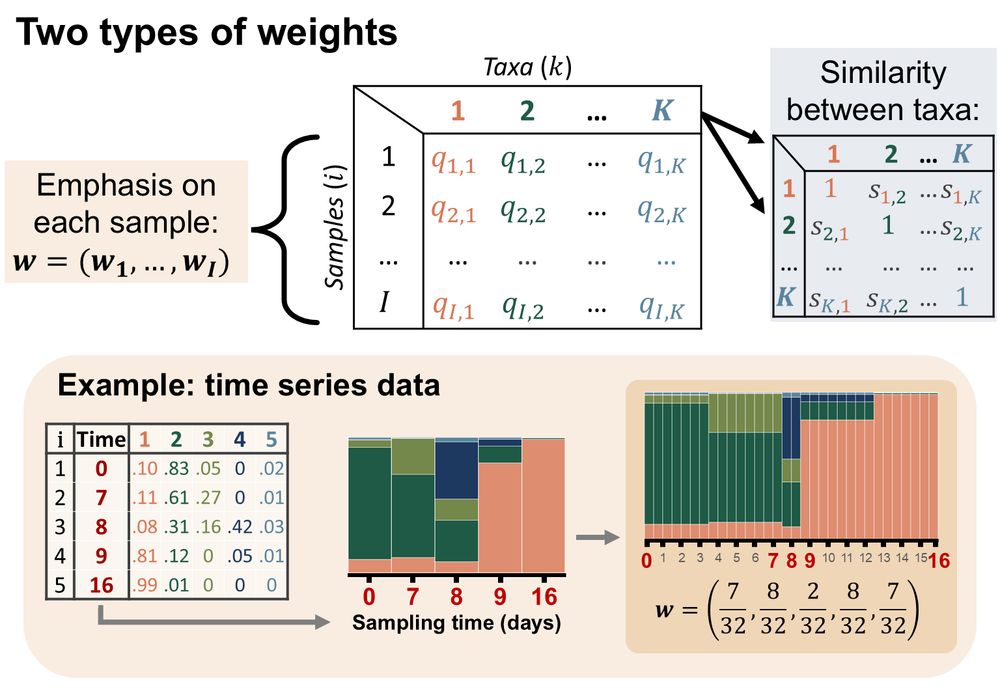
Diagram showing that FAVA can incorporate a matrix giving the similarity between each pair of taxa, as well as a vector of weights for each sample, which can be particularly useful for time series data with uneven sampling times.
5/ FAVA can account for phylogenetic similarity among taxa and can differentially weight samples, which is especially useful for time series data (Fig. 3). FAVA can be computed in sliding windows to see how variability changes over space or time (Fig. 4). We demonstrate FAVA in 2 examples.
14.03.2025 20:46 — 👍 5 🔁 0 💬 2 📌 04/ FAVA can analyze any relative abundance data, including taxonomic categories like OTUs and species, and functional categories like CAZymes and COGs. You can directly compare FAVA values computed on data with different numbers of categories (e.g., variability of bacterial species vs. families).
14.03.2025 20:46 — 👍 3 🔁 0 💬 1 📌 0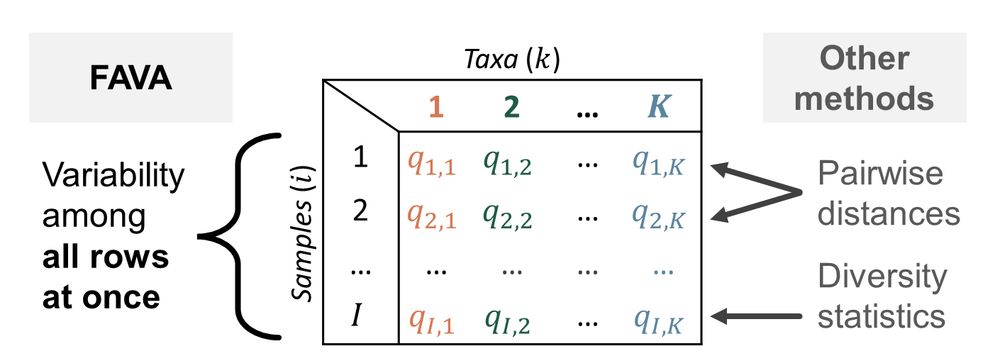
Schematic showing that FAVA measures variability across all rows of an OTU table at once, while other methods measure the diversity of one sample or the distance between a pair of samples.
3/ FAVA extends the pop-gen statistic FST (which traditionally measures variability of allele frequencies across multiple populations) to instead measure variability of taxonomic or functional abundances across multiple microbiome samples (Fig. 1). All it needs is an OTU table, or similar.
14.03.2025 20:46 — 👍 6 🔁 2 💬 1 📌 02/ FAVA is implemented in an R package available for download from CRAN (just run install.packages("FAVA") in R). For detailed guidance on the analysis of microbiome data with FAVA, check out the tutorial on the package website: maikemorrison.github.io/FAVA/article...
14.03.2025 20:46 — 👍 6 🔁 0 💬 1 📌 0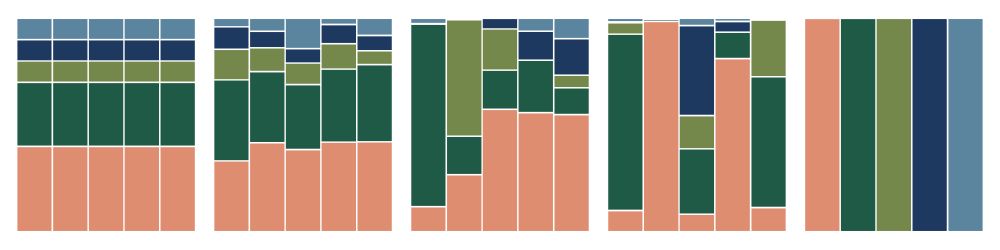
5 relative abundance plots arranged to have increasing compositional variability (variability across relative abundance samples, here vertical bars)
1/ Hey y'all, I'm excited to share my latest paper, which is out now in PNAS! We introduce FAVA, a statistical framework to measure compositional variability across microbiome samples. If you want to measure variability across a stacked bar plot, FAVA is for you! Paper: doi.org/10.1073/pnas...
14.03.2025 20:46 — 👍 207 🔁 72 💬 5 📌 26/ (Ex. 1) In microbiome samples collected along the GI tracts of individuals from 7 ruminant species, FAVA peaks in the small intestine–so lots of variation would have been missed by stool sampling! We also find taxonomic variability >> functional variability, evidence for functional redundancy!
14.03.2025 19:58 — 👍 0 🔁 0 💬 0 📌 05/ FAVA can account for phylogenetic similarity among taxa and can differentially weight samples, which is especially useful for time-series data (Fig. 3). FAVA can be computed in sliding windows to see how variability changes over space or time (Fig. 4). We demonstrate FAVA in 2 examples:
14.03.2025 19:58 — 👍 0 🔁 0 💬 1 📌 04/ FAVA can analyze any relative abundance data, including taxonomic categories like OTUs and species, and functional categories like CAZymes and COGs. You can directly compare FAVA values computed on data with different numbers of categories (e.g., variability of bacterial species vs. families).
14.03.2025 19:58 — 👍 0 🔁 0 💬 1 📌 03/ FAVA extends the pop-gen statistic FST (which traditionally measures variability of allele frequencies across multiple populations) to instead measure variability of taxonomic or functional abundances across multiple microbiome samples (Fig. 1). All it needs is an OTU table, or similar.
14.03.2025 19:58 — 👍 0 🔁 0 💬 1 📌 0I've been tremendously fortunate to work with @ksxue.bsky.social during her time as a postdoc at Stanford. She's not only a brilliant scientist and an excellent mentor/collaborator, but also a wonderful human being. You should totally join her lab!
14.03.2025 00:28 — 👍 6 🔁 0 💬 0 📌 0I’m thrilled to share my first ever publication, now published in PNAS! www.pnas.org/doi/10.1073/...
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
Reading "The Science Bargain" by Rush Holt: "Democracy is at risk when it becomes simply a contest of opinions not grounded in evidence. When one opinion is as good as another–each asserted as strongly or even as deceptively as possible–democracy cannot survive. This is a call to science."
12.03.2025 14:07 — 👍 0 🔁 0 💬 0 📌 0
NIH to ax grants on vaccine hesitancy, mRNA vaccines. www.science.org/content/arti...
10.03.2025 23:25 — 👍 218 🔁 124 💬 15 📌 54
Maike holding a sign in front of SF city hall. Sign is a parody of the Gadsden flag. Text on bottom says, "don't tread on me." Snake usually on Gadsden flag is replaced with a caterpillar phylogenetic tree.

Large crowd in front of SF city hall.
It's been easy to feel powerless these past few months, but the @standupforscience.bsky.social rally in SF this afternoon was an important reminder that the fight is not over!
08.03.2025 01:59 — 👍 7 🔁 0 💬 1 📌 0Today's small joy was discovering the latex package "etaremune," which you can use in place of "enumerate" to count in reverse
23.07.2024 21:54 — 👍 3 🔁 0 💬 0 📌 0
Beach with palm trees and a sailboat
I'm stoked to be at #SMBE2024! If you're interested in variability/diversity, microbiomes, and/or non-traditional applications of FST, I would love to chat!
Poster Weds #44, title: Quantifying the stability of microbiomes and the timescale of antibiotic perturbation with FAVA