 31.05.2025 01:20 — 👍 21220 🔁 4065 💬 417 📌 158
31.05.2025 01:20 — 👍 21220 🔁 4065 💬 417 📌 158
@dalygene.bsky.social
Geneticist…trying to make what we do useful for medicine. Blessed with amazing family. Hope you have a beautiful day!
@dalygene.bsky.social
Geneticist…trying to make what we do useful for medicine. Blessed with amazing family. Hope you have a beautiful day!
 31.05.2025 01:20 — 👍 21220 🔁 4065 💬 417 📌 158
31.05.2025 01:20 — 👍 21220 🔁 4065 💬 417 📌 158
Maybe corny but it was helpful for me to hear Henry Rollins saying "this is not time to be dismayed, this is punk rock time, this is what Joe Strummer trained you for... You can be thunderous in your own life, to the eight people around you. That rubs off... Goodness is viral"
23.02.2025 23:54 — 👍 16066 🔁 3135 💬 204 📌 195
Screenshot from "Letter from our correspondent" in "Papua New Guinea Courier"
The Papua New Guinea Courier is in rare form.
06.02.2025 12:31 — 👍 30024 🔁 10889 💬 1166 📌 1807Last call! The deadline for applications to our FIMM-EMBL International PhD Programme is tomorrow, 7 Feb!
Don't miss this opportunity to pursue your doctoral studies in a unique, ambitious and interdisciplinary research environment.
🔗 jobs.helsinki.fi/job/Helsinki...
#PhDposition #PhDopportunity
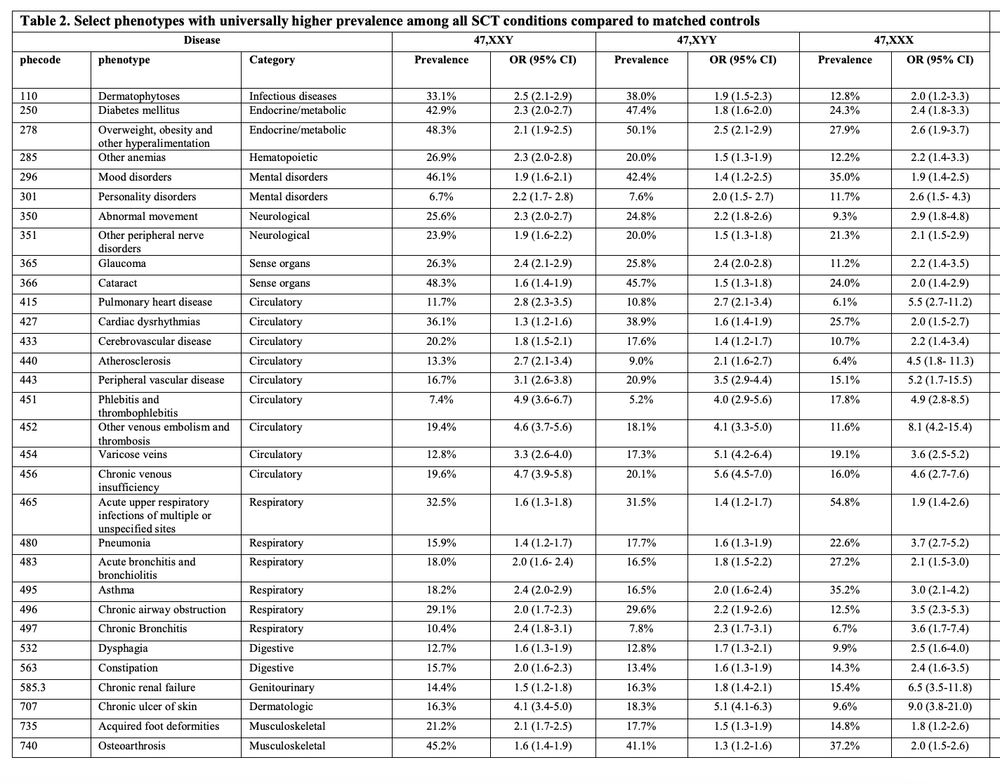
Nice paper showing ~1/540 people have a sex chromosome trisomy from 3 large biobanks (UKB, FinnGen, MVP) comprising 1.5 million individuals along with their phenotypic impact.
www.medrxiv.org/content/10.1...
 20.01.2025 21:51 — 👍 9757 🔁 2240 💬 192 📌 201
20.01.2025 21:51 — 👍 9757 🔁 2240 💬 192 📌 201
Curiously, the longer-standing very common signal at the locus that @timfrayling.bsky.social highlights has pinpointed the gene for years (and seems to include another missense variant in credible set rs10305420) seems more strongly associated to obesity in UKBB & FG...
10.01.2025 16:34 — 👍 2 🔁 0 💬 0 📌 0As @kauralasoo.bsky.social points out rs10305492 (A316T) is a lights out conclusively fine-mapped (PIP>0.99) for lower glucose and HbA1c in UKBB & FinnGen - T2D protective in multi-biobank mvp-ukbb.finngen.fi/variant/6:39...
10.01.2025 16:30 — 👍 6 🔁 2 💬 1 📌 0
Million Veteran Program & FinnGen teams are pleased to release v1 meta-analysis of MVP, FinnGen and UKBB GWAS data. This first version includes ~300 binary disease definitions across >1.5 M individuals.
Browse scans at: mvp-ukbb.finngen.fi

Independently reported a few days later by @alnylam.bsky.social in UKBB exomes: www.medrxiv.org/content/10.1...
27.11.2024 11:55 — 👍 3 🔁 0 💬 2 📌 0Sometimes going 5 levels of conditional fine-mapping reveals something new about an old association! Great work from @heikorunz.bsky.social @mpreeve.bsky.social Seppo Meri's Lab and @finngen.bsky.social
27.11.2024 11:53 — 👍 1 🔁 0 💬 1 📌 0Totally agree - FM from meta of heterogeneous GWAS is very imprecise (see @masakanai.bsky.social papers) and I cannot imagine anyone would say we have a near complete annotation of functional non-coding elements. Impossible to interpret then when FM points to unannotated variants...
22.11.2024 15:58 — 👍 1 🔁 0 💬 0 📌 0I agree with this, and would add that data of regulatory annotations doesn't cover all cell types where the variant effect might be driving the trait association. scATAC probably has the best coverage of cell types, but it's not perfect either.
20.11.2024 11:22 — 👍 11 🔁 1 💬 1 📌 0