
[4/4] Our approach is by design a meta-method: you can use it with your favorite single-cell RNA-based GRN inference tool, and squeeze more insights out of your data! Check us out GitHub: github.com/aliaaz99/GRN....
#singlecell #GRN #LLM
@nsapoval.bsky.social
Lost in between mathematics, computer science and biology. Hoping to find myself one day. https://nsapoval.github.io/

[4/4] Our approach is by design a meta-method: you can use it with your favorite single-cell RNA-based GRN inference tool, and squeeze more insights out of your data! Check us out GitHub: github.com/aliaaz99/GRN....
#singlecell #GRN #LLM
[3/4] We use these embeddings to construct a prior graph and then further refine it with some known TF-target interactions as pre-training targets. Finally, we use this augmented prior graph jointly with a GRN inferred by *any* other method, in order to produce a final prediction.
28.11.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
[2/4] We use gene descriptions from NCBI Gene database and embed them into a high-dimensional space with a LLM (Qwen3-8B). This idea was inspired by GenePT (pubmed.ncbi.nlm.nih.gov/37905130/) and a great study on gene embeddings from @vyao.bsky.social's group (www.biorxiv.org/content/10.1...).
28.11.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
Check out our new preprint on improving gene regulatory network inference by incorporating a prior from plain-text gene descriptions. It's a simple idea, but we show that it proves to be quite powerful and adaptable. [1/4]
www.biorxiv.org/content/10.1...



Nicolae Sapoval @nsapoval.bsky.social presented "Theoretical and empirical performance of pseudo-likelihood- based Bayesian inference of species trees under the multispecies coalescent"
A fantastic theory talk, offering intuitive insights!
Paper: doi.org/10.1101/2025.01.28.635282
As the next step, we aim to develop rigorous corrections to the pseudo-likelihood-based credibility intervals in order to further improve scalability and applicability of Baeysian phylogenomic inference.
02.02.2025 00:11 — 👍 0 🔁 0 💬 0 📌 0In our work we explore suitability of pseudo-likelihood for Bayesian phylogenomic inference. We show that using pseudo-likelihood greatly reduces the computational burden of the Bayesian inference. However, the inferred credibility intervals are overconfident.
02.02.2025 00:11 — 👍 0 🔁 0 💬 1 📌 0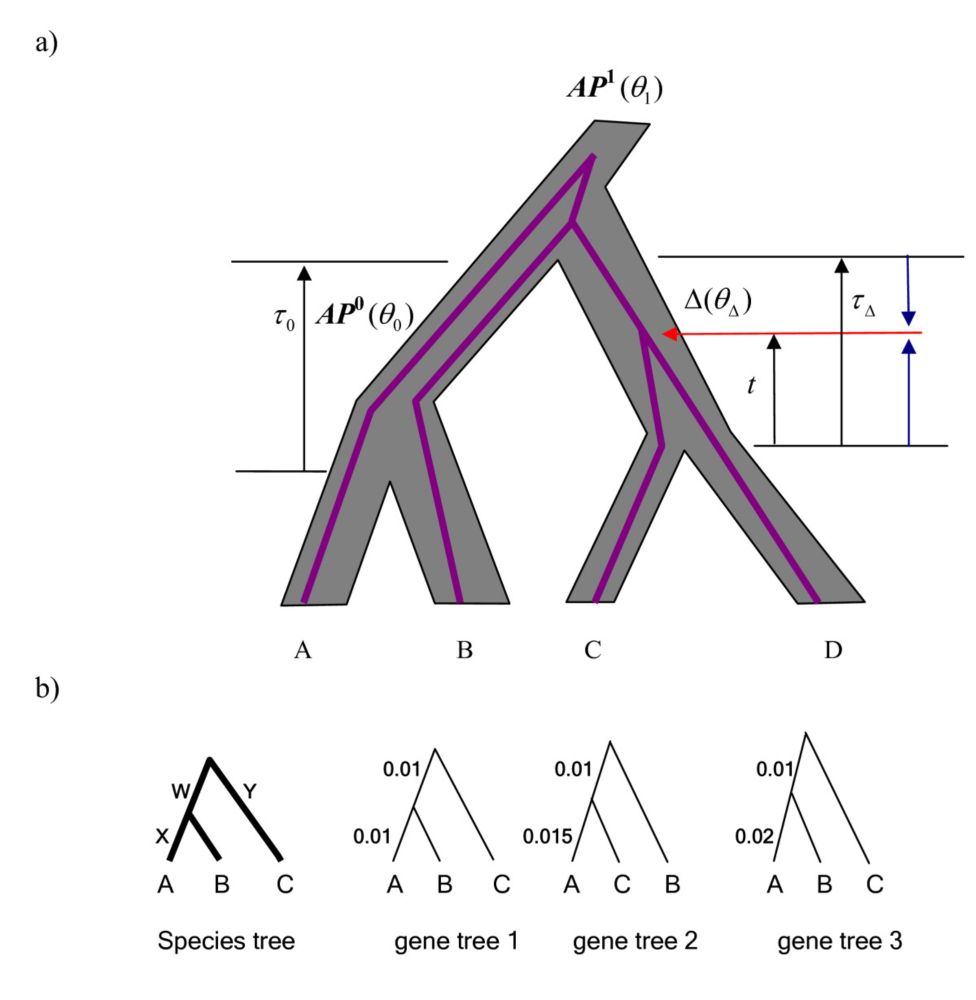
Likelihood-based phylogenomic inference is common, but it faces scalability issues. Hence, pseudo-likelihood has been previously proposed as a statistically consistent (for topology estimation) and scalable alternative: doi.org/10.1186/1471...
02.02.2025 00:11 — 👍 0 🔁 0 💬 1 📌 0
Our preprint on using pseudo-likelihood for Bayesian inference of species trees from gene tree data under the multispecies coalescent is now online: doi.org/10.1101/2025...
[1/n]
In case if you lose the URL (it’s not pretty), I have linked this on my website (nsapoval.github.io) as well.
01.01.2025 18:48 — 👍 1 🔁 0 💬 0 📌 0This is aimed primarily at the people who are just starting their thesis-based masters or a PhD. However, it’s also an evolving document, so suggestions and ideas are welcome!
01.01.2025 18:48 — 👍 1 🔁 0 💬 1 📌 0
It’s that time of the year when I get some writing done. Here are some notes on how I work with academic literature: plume-lifeboat-00b.notion.site/How-I-find-o...
01.01.2025 18:48 — 👍 1 🔁 1 💬 1 📌 0I just completed "Ceres Search" - Day 4 - Advent of Code 2024 #AdventOfCode adventofcode.com/2024/day/4 (A little bit behind the schedule this year, but gotta keep it going)
05.12.2024 19:19 — 👍 3 🔁 0 💬 0 📌 0
I was waiting for a great topic for my first Bluesky post, and I cannot thing of a better one: I’m thrilled to be hosting @aphillippy.bsky.social at Rice University today and looking forward to his talk at 4pm! events.rice.edu/event/345896...
14.11.2023 15:59 — 👍 13 🔁 2 💬 2 📌 0