Pre-assembly of biomolecular condensate seeds drives RSV replication
Nature - Viral ribonucleoprotein–viral protein networks form pre-replication centres that nucleate viral factories and drive respiratory syncytial virus replication.
Now out in Nature! We visualize infection of the RNA virus RSV in real-time with single-vRNP resolution to understand how RSV establishes viral factories, biomolecular condensates that act as sites of viral replication. A huge collaborative effort led by Dhanushika Ratnayake!
rdcu.be/e1bBW
28.01.2026 20:38 —
👍 90
🔁 35
💬 1
📌 2

Image with overview of Hubrecht Symposium 2026 information, including title, speakers, etc.
There are still spots available for our Hubrecht Symposium 2026, taking place on Thursday March 26!
📝 Abstract submission deadline: February 1st.
🎟️ Registration deadline: February 15th.
Learn more and register 👇
www.hubrecht.eu/hubrecht-sym...
26.01.2026 10:27 —
👍 6
🔁 2
💬 0
📌 1
So many exciting things ahead, Robin. Enjoy it all!
02.01.2026 15:37 —
👍 3
🔁 1
💬 0
📌 1

🌱 As of Jan 1, I started as Assistant Professor at UMC Utrecht – Center for Molecular Medicine.
Very grateful to @dewitlab.bsky.social and @jopkind.bsky.social and to the many other people who supported me along the way.
I’ll be launching the lab soon, focusing on Chromatin Systems Biology.
02.01.2026 13:56 —
👍 29
🔁 4
💬 3
📌 2
Group Leader - Generative Biology and AI
Do you want to help us improve human health and understand life on Earth? Make your mark by shaping the future to enable or deliver life-changing science to solve some of humanity’s greatest challenge...
Looking to start your lab in generative biology / AI?
Come join us at the @sangerinstitute.bsky.social
Sanger is core-funded so you can generate data at scale to train the next generation of models and understanding. Design/Engineering/Chemistry/Proteins/Pathways!
pls RT
tinyurl.com/GenGenFaculty
01.01.2026 12:08 —
👍 33
🔁 33
💬 0
📌 0

1/
🧬 Very excited to share our work on dissecting imputation strategies for single-cell histone modification data! academic.oup.com/nargab/artic...
A huge thank you to @robinhweide.bsky.social, @jopkind.bsky.social and Jeroen de Ridder for their collaboration in this project!
30.12.2025 14:29 —
👍 10
🔁 3
💬 1
📌 1
6/ Together, our work links nuclear architecture, epigenetic state, and MeCP2 activity to the spatiotemporal control of long neuronal genes.
With implications for understanding Rett syndrome and broader mechanisms of cortical development.
Great collaboration with Basak (UMC Utrecht) team
🧵//
17.09.2025 08:48 —
👍 0
🔁 0
💬 0
📌 0
5/ Strikingly, MeCP2’s function appears context-dependent:
• In prenatal cortex: MeCP2 promotes activation of long genes.
• In postnatal cortex: MeCP2 represses the same genes
17.09.2025 08:46 —
👍 0
🔁 0
💬 1
📌 0
4/ Mechanistically, we identify MeCP2 (mutated in Rett syndrome) as a candidate regulator.
MeCP2 binds long LAD genes before they are released and transcribed, pointing to a stage-specific role in gene priming.
17.09.2025 08:45 —
👍 0
🔁 0
💬 1
📌 0
3/ These long neuronal genes often detach from the nuclear lamina before activation, suggesting lamina release is an early regulatory step in corticogenesis.
This spatial reorganization aligns with their role in neurodevelopmental disorders, including ASD.
17.09.2025 08:44 —
👍 0
🔁 0
💬 1
📌 0
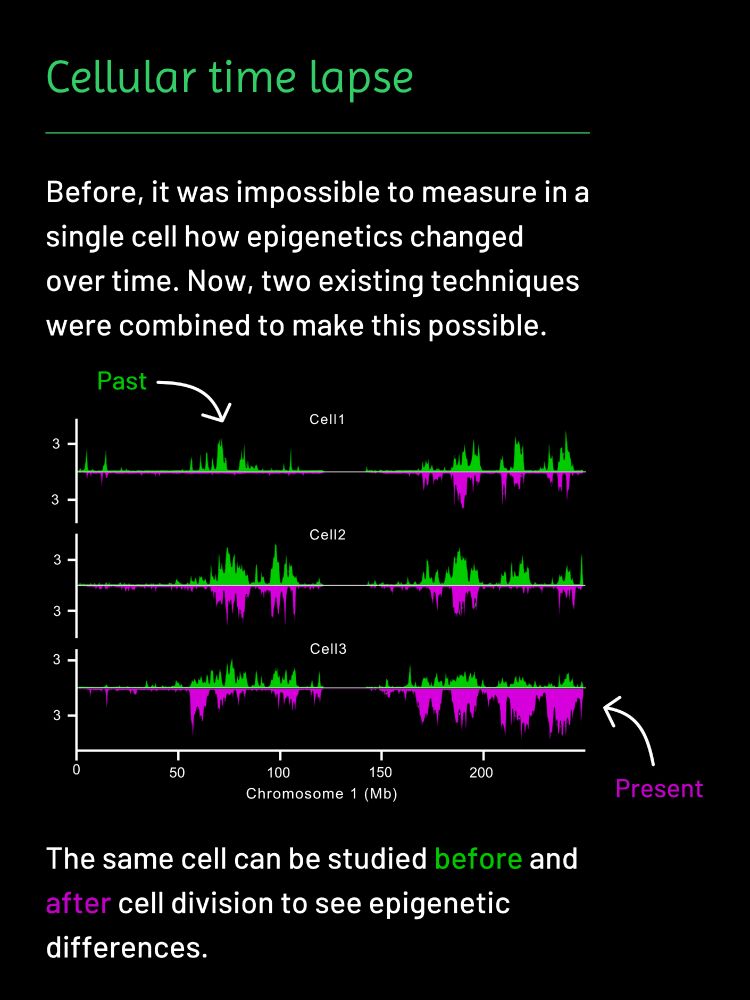
2/ Using in utero electroporation + scDam&T-seq, we jointly profiled transcriptomes and lamina-associated domains (LADs) in embryonic cortex.
We found widespread lamina reorganization during differentiation, strongly enriched at long neuronal genes (≥100 kb).
17.09.2025 08:42 —
👍 0
🔁 0
💬 1
📌 0

Join this year's CSH Asia Systems Biology of Gene Regulation & Genome Editing meeting in beautiful Suzhou, China, Oct 20-24. Let's bring the international communities together! Abstract deadline Sept 19! Infos and registration at csh-asia.org?content/2767. Please repost!
11.09.2025 06:09 —
👍 31
🔁 18
💬 2
📌 0
Very proud of Kim to be featured by MIT Technology Review as one of this year's innovators under 35 for her pioneering work on mapping DNA repair in single cells! @oncodeinstitute.bsky.social @hubrechtinstitute.bsky.social
11.09.2025 08:59 —
👍 11
🔁 1
💬 0
📌 0
This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇
18.08.2025 13:00 —
👍 113
🔁 34
💬 10
📌 0

Extensive differences observed in 3D genome structure between homologous heterozygous chromosomes revealed by Genome Architecture Mapping #GAM in the @apombo1.bsky.social lab ➡️ www.embopress.org/doi/full/10....
15.05.2025 17:32 —
👍 21
🔁 11
💬 1
📌 0

We’re looking for curious, innovative science leaders at EMBL Heidelberg! 🔬🧬🦠
Join a vibrant, interdisciplinary community where collaboration and innovation are nurtured at all levels.
Take a look at these four open positions 👇
07.08.2025 08:28 —
👍 73
🔁 73
💬 2
📌 4
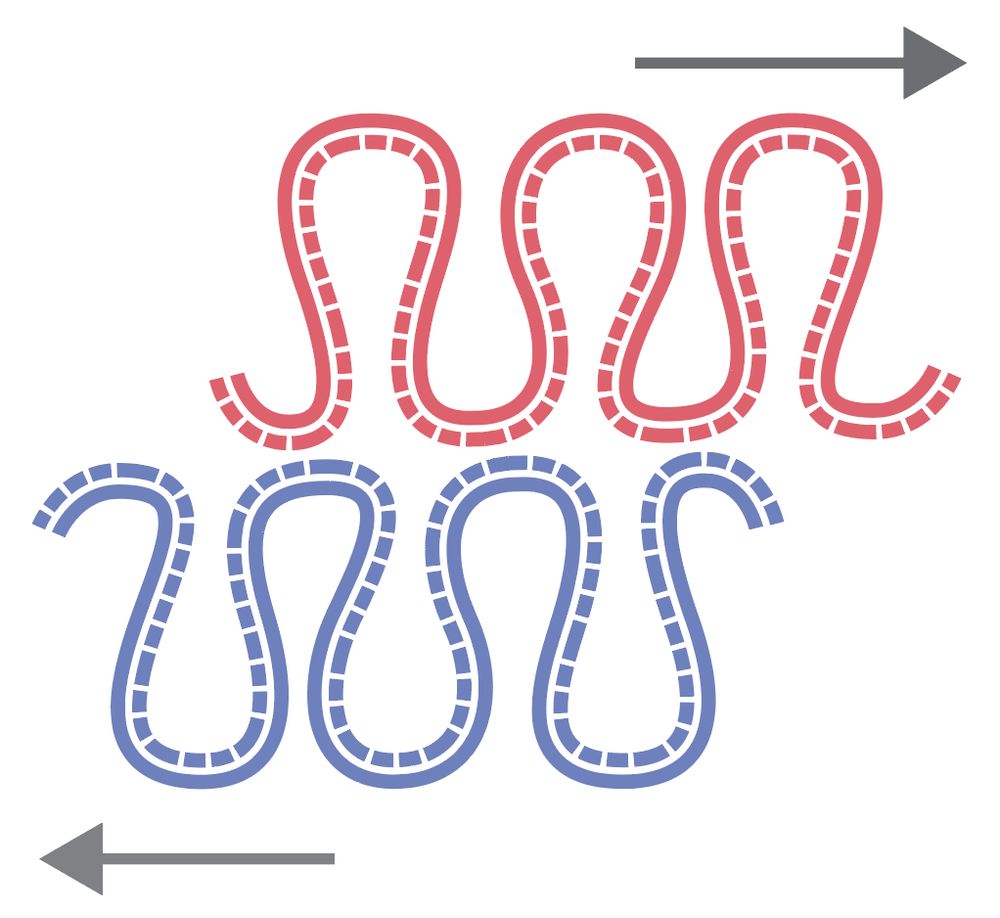
We found a new asymmetry in the large-scale chromosome structure: sister chromatids are systematically shifted by hundreds of kb in the 5′→3′ direction of their inherited strands! The work was led by Flavia Corsi, in close collaboration with the Daniel Gerlich lab.
www.biorxiv.org/content/10.1...
1/
15.07.2025 08:11 —
👍 115
🔁 59
💬 3
📌 7

🧬 Looking for a PhD position in molecular or developmental biology? 🔬
✉️ The Hubrecht Institute now has its very own PhD program! Applications are open and will be accepted until September 15th. Read more here 👉 www.hubrecht.eu/hipp/
10.07.2025 11:19 —
👍 14
🔁 13
💬 0
📌 5
Image-based, pooled phenotyping reveals multidimensional, disease-specific variant effects
Genetic variants often produce complex phenotypic effects that confound current assays and predictive models. We developed Variant in situ sequencing (VIS-seq), a pooled, image-based method that measures variant effects on molecular and cellular phenotypes in diverse cell types. Applying VIS-seq to ~3,000 LMNA and PTEN variants yielded high-dimensional morphological profiles that captured variant-driven changes in protein abundance, localization, activity and cell architecture. We identified gain-of-function LMNA variants that reshape the nucleus and autism-associated PTEN variants that mislocalize. Morphological profiles predicted variant pathogenicity with near-perfect accuracy and distinguished autism-linked from tumor syndrome-linked PTEN variants. Most variants impacted a multidimensional continuum of phenotypes not recapitulated by any single functional readout. By linking protein variation to cell images at scale, we illuminate how variant effects cascade from molecular to subcellular to cell morphological phenotypes, providing a framework for resolving the complexity of variant function. ### Competing Interest Statement FPR is an advisor and shareholder in Constantiam Biosciences. National Human Genome Research Institute, https://ror.org/00baak391, RM1HG010461, R01HG013025 National Institute of General Medical Sciences, R35GM152106 National Heart Lung and Blood Institute, https://ror.org/012pb6c26, K99HL177347, R01HL171174, R01HL164675 Chan Zuckerberg Initiative (United States), CZIF2024-010284, CP-2-1-Fowler Brotman Baty Institute, https://ror.org/03jxvbk42, CC28 United States Department of Veterans Affairs, I01BX006428, IK2BX004642 Novo Nordisk Foundation, https://ror.org/04txyc737, Alex's Lemonade Stand for Childhood Cancer RUNX1 Foundation
⚡ I developed VIS-seq with the help of dougfowler.bsky.social and others at UW Genome Sciences. Check out my preprint:
www.biorxiv.org/content/10.1...
2/9
07.07.2025 02:44 —
👍 4
🔁 1
💬 1
📌 0

UMAP of LMNA variant profiles (left) colored by Louvain-derived clusters. Colors legend at bottom.
Graphical representation (right) of lamin A processing, localization, aggregation, and degradation, annotated with their relationship to dimerization, multimerization and variant clusters.
🌎 LMNA variant ➡️ structure ➡️ abundance and localization ➡️ function! VIS-seq maps LMNA variant effects across scales of cellular organization and discovered a new subset of gain-of-function LMNA variants.
7/9
07.07.2025 02:44 —
👍 2
🔁 1
💬 1
📌 0
Exciting opportunity at @scilifelab.se
04.07.2025 07:22 —
👍 2
🔁 2
💬 0
📌 0
Beautiful work Grace!! Congratulations!
03.07.2025 19:26 —
👍 0
🔁 0
💬 0
📌 0

Range extender mediates long-distance enhancer activity - Nature
The REX element is associated with long-range enhancer–promoter interactions.
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 —
👍 186
🔁 90
💬 10
📌 9

Delighted that our work on positional memory is now published. We asked how axolotl cells 'know' which part of the limb to regenerate after injury.
www.nature.com/articles/s41...
A joy to work with super team Sarah Plattner, Yuka Sugiura, Francisco Falcon and Elly Tanaka.
🧵1/14
21.05.2025 15:05 —
👍 211
🔁 66
💬 23
📌 9

Thesis cover image
🧬What makes one cell different from another? Kim de Luca developed methods that help answer this question. They can be used to study DNA organization, regulation and repair in single cells. Today she successfully defended her thesis. Congratulations Kim!🎓 Read more 👉 www.hubrecht.eu/phd-kim-de-l...
30.06.2025 14:46 —
👍 7
🔁 1
💬 0
📌 0
Today, on ‘Be Open about Animal Research Day’, we spotlight how research with zebrafish brings us closer to healthier hearts. #BOARD25
Sharing about 80% of heart disease–causing genes with humans, the zebrafish is an excellent model to study how hearts develop, become diseased and might regenerate.
03.06.2025 08:34 —
👍 9
🔁 3
💬 1
📌 0