PHLAME is available to try out on GitHub:
github.com/quevan/phlame
@quevan.bsky.social
PhD candidate in the MIT Microbiology Program (co2020). @Lieberman Lab @contaminatedsci.bsky.social. I study the ecology and evolution of skin microbes, like this one -> 🤏

PHLAME is available to try out on GitHub:
github.com/quevan/phlame
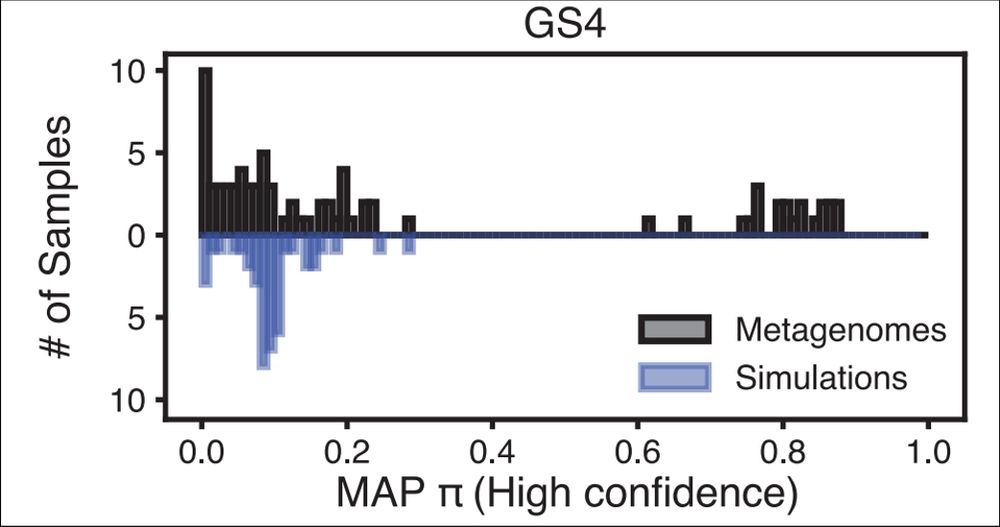
Turning to the vaginal microbiome, we showed how PHLAME's novelty aware approach can identify samples that are abundant novel diversity.
By our estimate, about 1/3 of vaginal samples we looked at had substantial abundances (>20%) of yet-characterized Gardnerella strains

We also found a clade of C. acnes that is higher abundance on older people (>40 yo). This association is independent of sex and consistent across geographic regions.
11.02.2025 23:06 — 👍 1 🔁 0 💬 1 📌 0
We used PHLAME to pull out some interesting associations from public data.
In the skin microbiome, we discovered that some clades of C. acnes are recently emerged, strongly geographic restricted, and at high prevalence in those regions. This pattern may indicate region specific adaptation.

Using this novelty-aware approach, PHLAME achieves near-perfect precision and high sensitivity, even for species with low coverage.
We also benchmarked PHLAME using @microjacob.bsky.social's unique resource of thousands of paired isolates and metagenomes from the same samples (see Fig. 4)

Counting missing mutations is not easy when reads are low-coverage and overdispersed.
We solved this problem using a model that independently measures dispersion and zero-inflation by comparing counts across just the mutational allele compared to all alleles at the same positions.

We figured out that we could estimate the divergence along each branch (novelty of a new strain) by counting the proportion of clade-specific mutations missing in metagenomic samples (π).
11.02.2025 23:06 — 👍 1 🔁 0 💬 1 📌 0
PHLAME quantifies novel strain diversity in samples using an evolutionary framework. Novel strains in a sample are assumed to share some, but not all, evolutionary history with known strains. We call the degree of unshared evolutionary history between a sample and a reference database Divergence.
11.02.2025 23:06 — 👍 1 🔁 0 💬 1 📌 0
We were concerned that ‘pushing’ novel diversity might influence downstream association detection, especially in samples with significant amounts of novel strain diversity.
11.02.2025 23:06 — 👍 1 🔁 0 💬 1 📌 0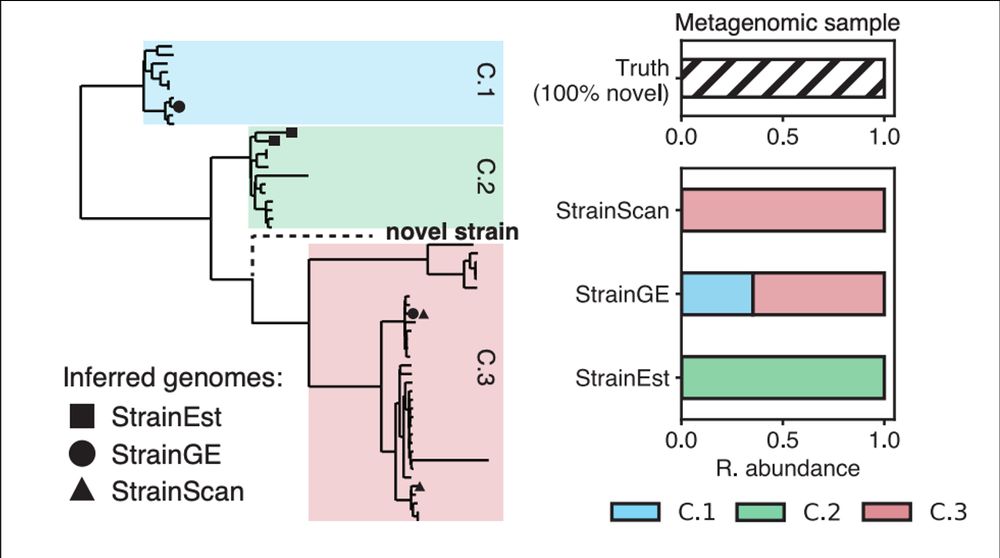
Standard practice for complex metagenomic samples is to use reference databases to detect strains.
Because reference databases are never comprehensive, many of these methods will represent novel strains (i.e., not in the database) as a nearby representative genome in the database.
Many health or environmental associations may be driven by intraspecies variants.
The most straightforward approach for strain associations, direct inference of genotypes from metagenomics, is difficult in environments where many strains of the same species coexist.

Excited to share my preprint describing a new microbiome analysis method, PHLAME, for detecting strain-level associations in difficult sample types. 🧵
www.biorxiv.org/content/10.1...
This work was done with my great coauthors and my amazing advisor @contaminatedsci.bsky.social
Ever wondered about the origin of the bacteria that call our faces home? 🤔 Our new preprint dives into the fascinating dynamics of the human facial skin microbiome (FSM) and explores the natural history of important microbiome species on people at high resolution. 🧫🧵
11.01.2024 17:48 — 👍 23 🔁 12 💬 1 📌 2