We are on the lookout for postdocs for two different projects at the intersection of ecology, evolution, and the human microbiome.
See thread for more information and reach out!
Laura Markey
@lauramarkey.bsky.social
@lauramarkey.bsky.social
We are on the lookout for postdocs for two different projects at the intersection of ecology, evolution, and the human microbiome.
See thread for more information and reach out!
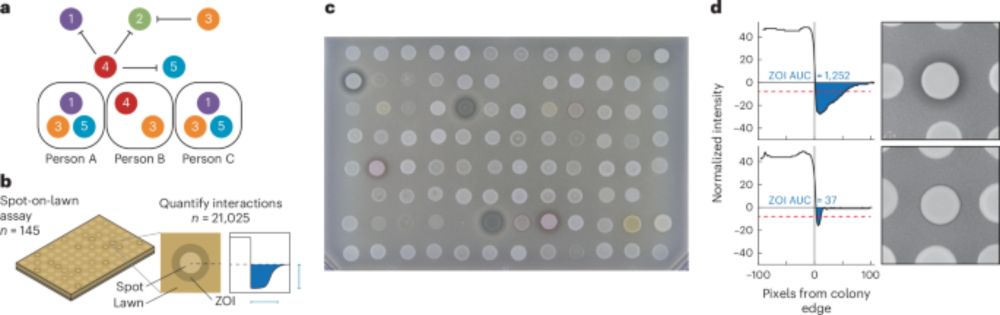
🚨Out now!
About 15,000 pairwise interactions within S. epidermidis from 18 people in 6 families reveal the antagonism and molecular trade-offs that shape the skin microbiota @contaminatedsci.bsky.social @mancusosci.bsky.social #MicrobiomeSky
www.nature.com/articles/s41...
read here: rdcu.be/et7VP
Thank you!!
25.06.2025 15:46 — 👍 1 🔁 0 💬 0 📌 0Thanks to labmate and co-author @quevan.bsky.social , additional co-authors and my PI @contaminatedsci.bsky.social ! 4/4
25.06.2025 12:29 — 👍 3 🔁 0 💬 1 📌 0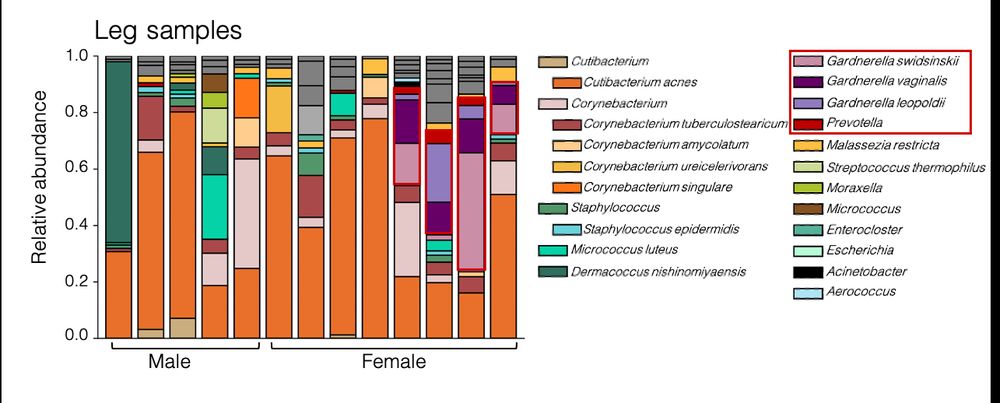
barplot of individual leg skin microbiome profiles demonstrates diverse composition across subjects including multiple species of Cutibacterium, Corynebacterium and Staphylococcus. 4/8 female subjects have multiple vaginal microbiome species including Gardnerella and Prevotella but not Lactobacillus present at >10% abundance.
Metagenomics enabled the surprising observation that common vaginal microbiome member Gardnerella (but not Lactobacillus) made up at >10% of the low biomass lower leg skin microbiome of 4/8 female subjects. 3/4
25.06.2025 12:29 — 👍 3 🔁 0 💬 1 📌 0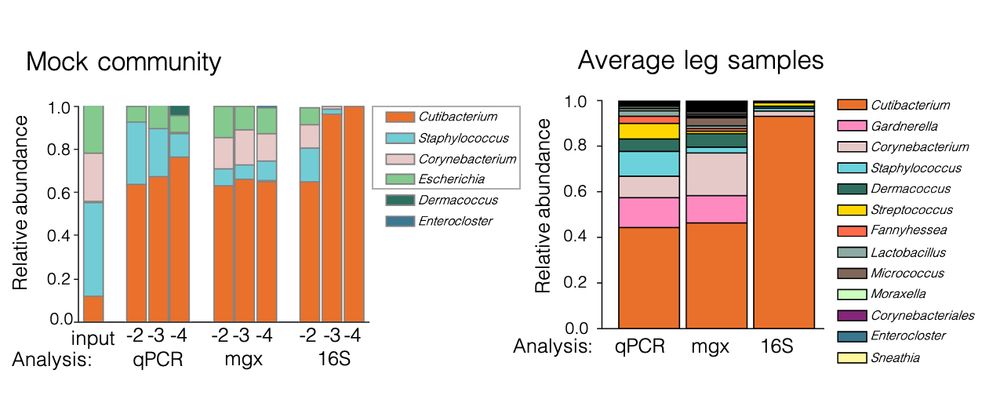
Left barplot shows that as mock community microbiome sample is diluted, qPCR and metagenomics continue to detect all taxa but 16S detects only Cutibacterium. Right barplot shows that for qPCR and metagenomics detect a diverse average microbiome from lower leg skin but 16S composition is dominated by Cutibacterium.
In high biomass samples, sequencing methods are comparable – however, as biomass decreases, minor taxa are rapidly lost with 16S as the limited amplicon library becomes biased towards the most abundant taxon. 2/4
25.06.2025 12:29 — 👍 6 🔁 3 💬 1 📌 0
Excited to share my new preprint: we show that for lab control and skin samples with low microbial biomass, metagenomics significantly outperforms 16S while DNA extraction method has minimal impact. 🧵 1/4
www.biorxiv.org/content/10.1...
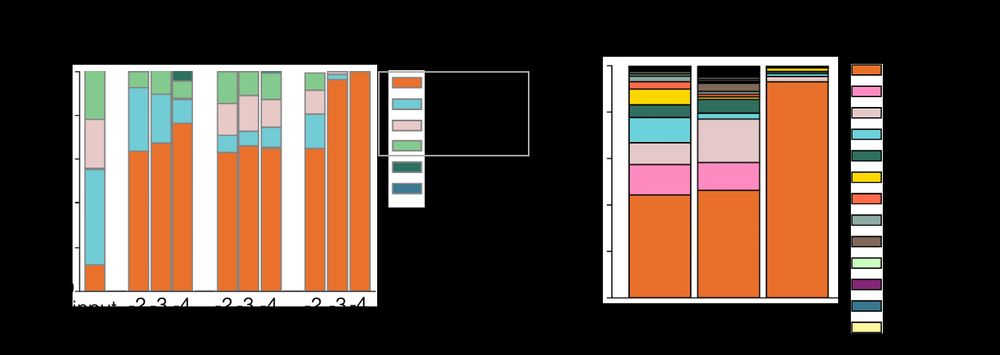
Left side of image shows a barplot of mock microbiome sample dilution series analyzed by qPCR, metagenomics and 16S sequencing- as the sample is diluted, qPCR and metagenomics continue to recall all taxa while 16S is increasingly dominated by Cutibacterium. Right side of image shows that similarly qPCR and metagenomics detect a diverse average leg skin microbiome while 16S is dominated by Cutibacterium.
In high biomass samples, sequencing methods are comparable – however, as biomass decreases, minor taxa are rapidly lost with 16S as the limited amplicon library becomes biased towards the most abundant taxon. 2/4
25.06.2025 12:22 — 👍 0 🔁 0 💬 0 📌 0
Our work on the facial skin microbiome of non-human primates is out in mSystems!
We show there is no close relative of Cutibacterium on the faces of gorillas and chimps at the Lincoln Park Zoo, furthering the mysterious origin of the dominant human skin colonizer.
journals.asm.org/doi/10.1128/...
New pre-print from my grad thesis lab! So excited to see this interesting paper for itself and to see that the gut-Candida-brain arm of the Kumamoto lab continues on
28.02.2025 02:40 — 👍 1 🔁 0 💬 0 📌 0Staph is so sticky!! Once thought I had suddenly forgotten how to make dilutions when CFU plating (for an OD/CFU curve) reps were terrible - nope just so sticky I had to use washed culture to get CFUs that were actually single cells dropping onto a plate
27.02.2025 01:08 — 👍 1 🔁 0 💬 1 📌 0
Use this form to tell us how the new US administration is affecting your research, or suggest future coverage
https://go.nature.com/4aYn9Me

Excited to share my preprint describing a new microbiome analysis method, PHLAME, for detecting strain-level associations in difficult sample types. 🧵
www.biorxiv.org/content/10.1...
This work was done with my great coauthors and my amazing advisor @contaminatedsci.bsky.social
👋 yeah I've been trying to "just move to industry" for a year so uh good luck with that
24.01.2025 18:13 — 👍 1 🔁 0 💬 1 📌 0
Candida albicans enhance Staphylococcus aureus virulence by progressive generation of new phenotypes https://www.biorxiv.org/content/10.1101/2024.06.26.600854v1
29.06.2024 17:16 — 👍 0 🔁 1 💬 0 📌 0
Siderophore piracy enables the nasal commensal Staphylococcus lugdunensis to antagonize the pathogen Staphylococcus aureus https://www.biorxiv.org/content/10.1101/2024.02.29.582731v1
02.03.2024 15:22 — 👍 3 🔁 1 💬 0 📌 0
Binning Singletons logo. A circle with 4 microbes. From left to right: a large pink vibrio on the left, smiling and pointing to the right; 4 yellow cocci looking around; a purple E. coli looking off to the right; and a green acetinobacter looking to the left. They represent the mentor and singletons at a large meeting and the mentor is helping them through all the facets of the meeting.
We're gearing up for #ASMicrobe in Atlanta! We help 1st-time and solo attendees have better experiences through networking and mentoring Here's a writeup on how it works
blog.addgene.org/binning-sing...
Ever wondered about the origin of the bacteria that call our faces home? 🤔 Our new preprint dives into the fascinating dynamics of the human facial skin microbiome (FSM) and explores the natural history of important microbiome species on people at high resolution. 🧫🧵
11.01.2024 17:48 — 👍 23 🔁 12 💬 1 📌 2
Highly-resolved within-species dynamics in the human facial skin microbiome https://www.biorxiv.org/content/10.1101/2024.01.10.575018v1
10.01.2024 22:19 — 👍 3 🔁 2 💬 0 📌 3Thanks! I'll have to check out what it's doing on cheese.
I don't know about most labs - but in our facility we can routinely recover S. xylosus both from the hairy, healthy skin of mice housed in the facility and from surfaces (lab benches, housing racks).
Thanks! I would add to what Tami said that we applied the bacteria (including E coli and L reuteri) every day - so even if they were at a disadvantage for survival on mouse skin, they were grown up in lab and reapplied daily and still did not delay healing
06.12.2023 02:09 — 👍 2 🔁 0 💬 1 📌 0 Bonus: Neither E. coli nor L. reuteri delayed healing, indicating this phenotype is limited to skin commensals!
Thanks to co-first Veda Khadka, and co-author Magalie Boucher, PI @contaminatedsci.bsky.social and Lieberman Lab for feedback! [3/3]
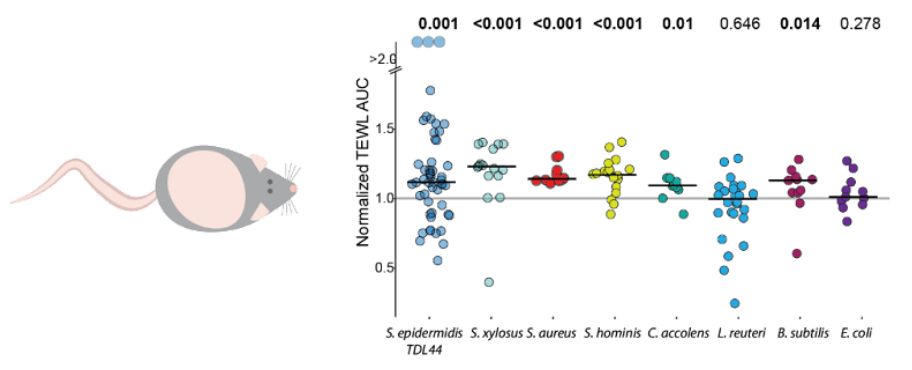
The answer: not when the skin is damaged! We tested multiple bacterial species, including mouse commensal S. xylosus, C. accolens, and 3 different isolates of S. epidermidis.All delayed healing when applied to abraded mouse flank skin. [2/3]
05.12.2023 19:26 — 👍 2 🔁 0 💬 2 📌 0Excited to announce a new preprint from the Lieberman Lab @contaminatedsci.bsky.social . We use a mouse model to ask: can we use commensal skin bacteria as topical probiotics? www.biorxiv.org/content/10.1... [1/3]
05.12.2023 19:21 — 👍 31 🔁 14 💬 1 📌 1