
Museum genomics reveals temporal genetic stasis and global genetic diversity in Arabidopsis thaliana
10.02.2025 20:53 — 👍 3 🔁 4 💬 0 📌 0
@ahmedelfarargi.bsky.social
Postdoc fellow @UT_AUSTIN | Evolutionary genomics | Local adaptation | Population genetics | Plant stress | Alumnus @MPIPZ_KÖLN @WUR

Museum genomics reveals temporal genetic stasis and global genetic diversity in Arabidopsis thaliana
10.02.2025 20:53 — 👍 3 🔁 4 💬 0 📌 0
Sweeps in space: leveraging geographic data to identify beneficial alleles in Anopheles gambiae
09.02.2025 16:54 — 👍 0 🔁 0 💬 0 📌 0
Quickdraws: a GWAS method to perform scalable association testing for quantiative and binary traits, without compromising computational efficiency
10.01.2025 10:25 — 👍 2 🔁 0 💬 0 📌 0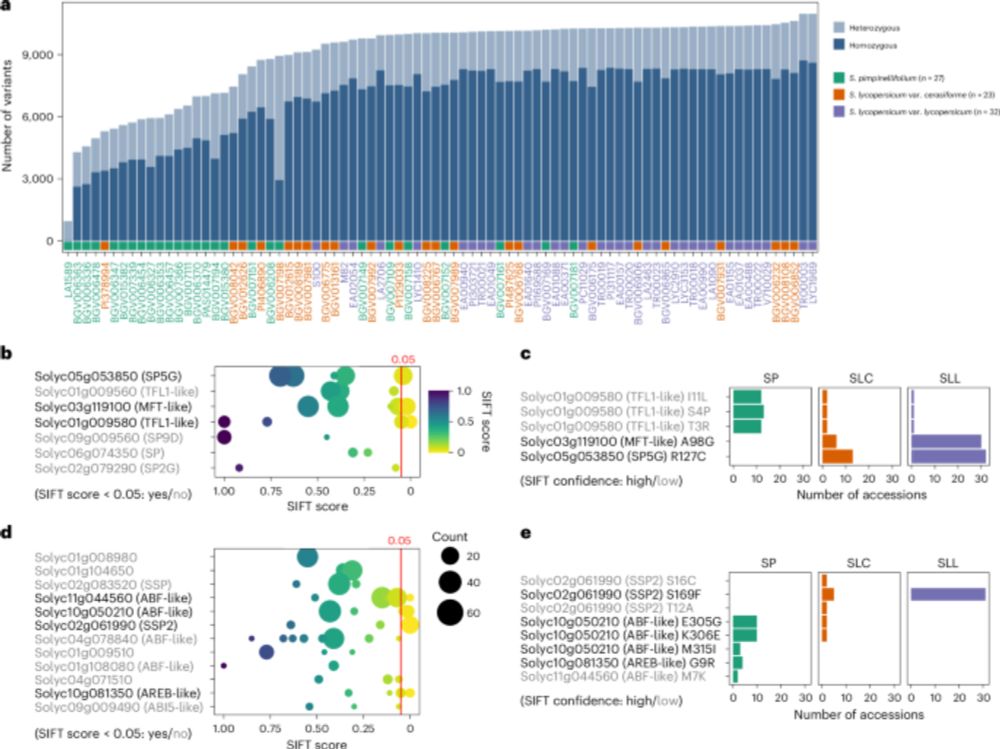
Very happy to see our work on deleterious mutations in #tomato published in @naturegenet.bsky.social 🧬🌱🍅! We identify deleterious mutations that were enriched during #domestication and apply #CRISPR to repair a mutation in cultivated tomato. #genetics #plantscience
www.nature.com/articles/s41...

A rare dominant allele DYSOC1 determines seed coat color and improves seed oil content in Brassica napus
06.01.2025 05:53 — 👍 1 🔁 0 💬 0 📌 0
A DNA language model based on multispecies alignment predicts the effects of genome-wide variants
06.01.2025 02:24 — 👍 0 🔁 0 💬 0 📌 0
The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum
06.01.2025 02:22 — 👍 1 🔁 2 💬 0 📌 0
RNAGenesis: Foundation Model for Enhanced RNA Sequence Generation and Structural Insights
06.01.2025 02:15 — 👍 0 🔁 0 💬 0 📌 0
Natural variation modifies centromere proximal meiotic crossover frequency and segregation distortion in Arabidopsis thaliana
05.01.2025 06:59 — 👍 1 🔁 0 💬 0 📌 0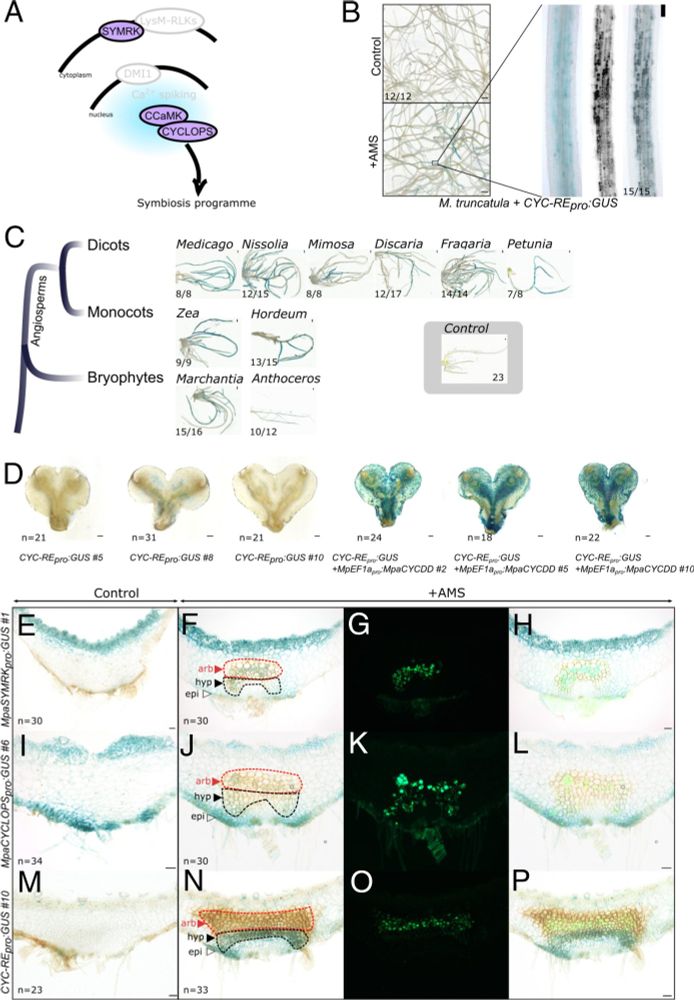
Really pleased to transition from 2024 to 2025 with Tatiana Vernie's work on the evolution of the common symbiosis pathway published @pnas.org 🔽
We finally demonstrate that ☘️ have maintained a genetic pathway to engage with🍄for half a billion years!
www.pnas.org/doi/10.1073/...
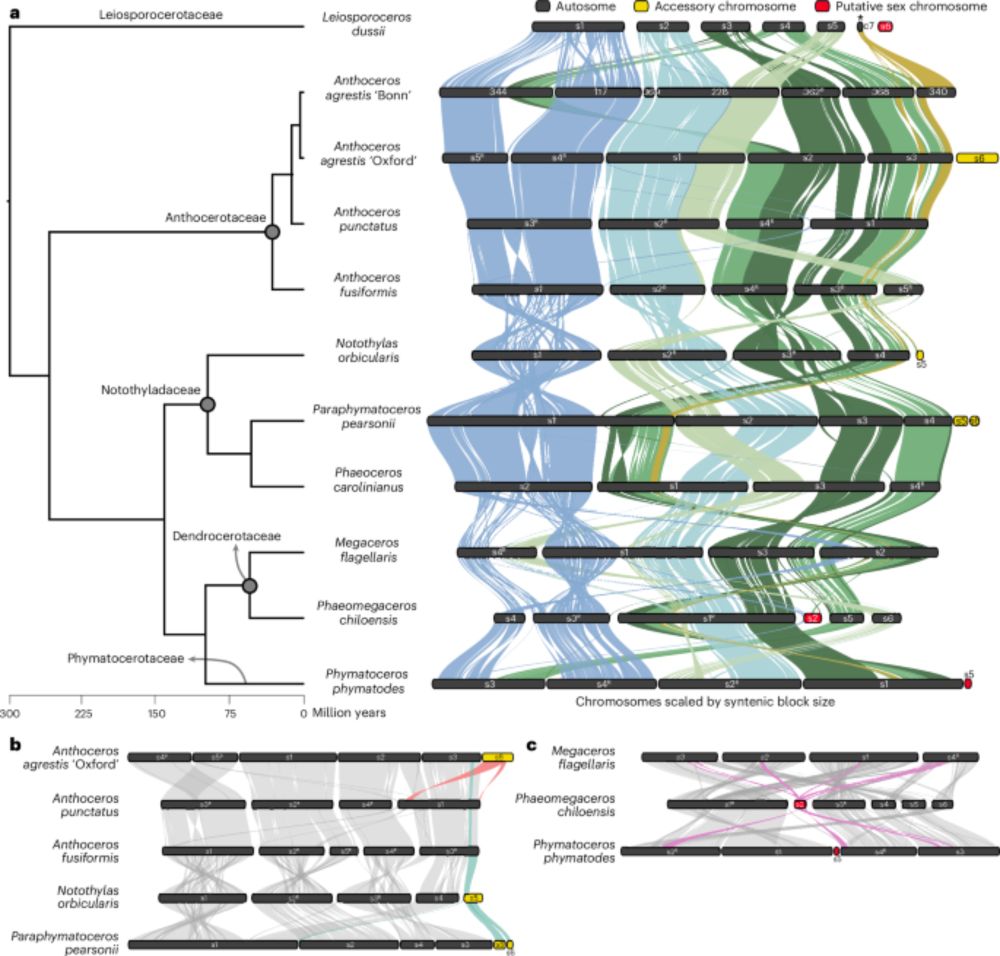
Paper #2 - we generated 10 new chr-level genomes for hornworts covering all families and most genera. Also the first look into their U/V sex chromosomes! 3/4 www.nature.com/articles/s41...
03.01.2025 11:11 — 👍 72 🔁 36 💬 1 📌 1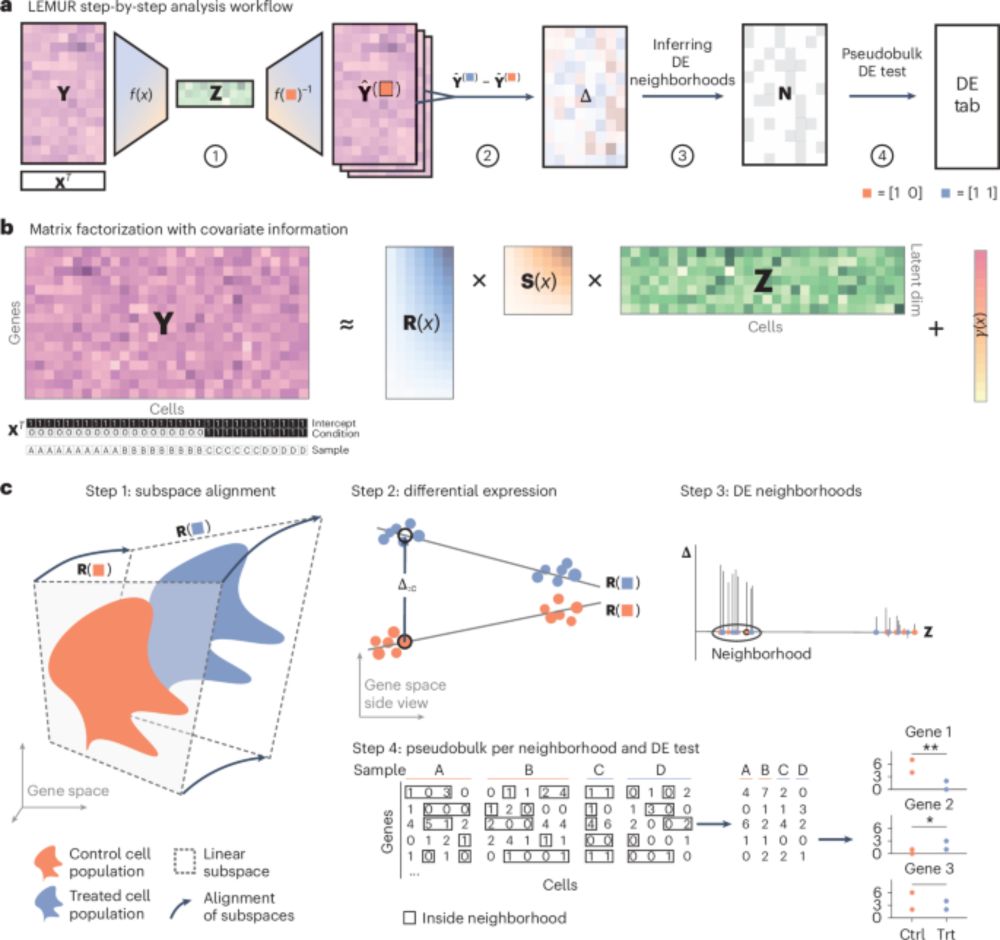
How to do differential expression with scRNAseq data? State of the art is "pseudo-bulk" analysis with RNA-seq methods like edgeR or DESeq2, where "cell type" is encoded as discrete categories. Biologically, discrete categories are not always the most appropriate concept.(1/3)
doi.org/10.1038/s415...

Increasing aridity may threaten the maintenance of a plant defence polymorphism
04.01.2025 19:49 — 👍 1 🔁 0 💬 0 📌 0Gabriele's paper is out in PNAS! He found a large number of genes with repeated sweeps in multiple plant species. Most interestingly: they tend to be peripheral in co-expression networks (low pleiotropy), consistent with mig-sel theory!! @gabnocgenomics.bsky.social www.pnas.org/doi/10.1073/...
02.01.2025 21:31 — 👍 24 🔁 11 💬 1 📌 2We may have cut our last Canu release in 2024, but we are starting off 2025 with Verkko2! 🎉 Not only is it 4x faster than Verkko1, this version integrates Hi-C data for both phasing and scaffolding, enabling the automated assembly of acrocentric chromosomes! Preprint: www.biorxiv.org/content/10.1...
02.01.2025 15:27 — 👍 86 🔁 31 💬 5 📌 1
Science magazine front cover. Picture of a walnut in its opened shell. The text reads "All about timing. Mating type evolution in walnuts and pecans"
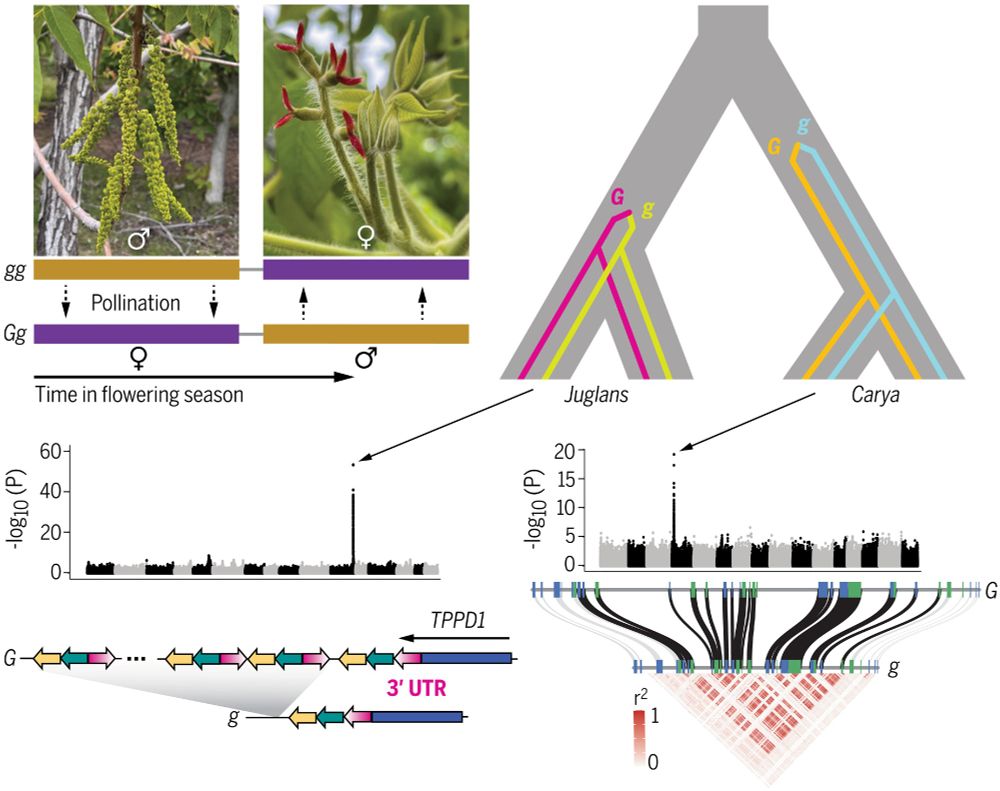
graphical abstract: "In Juglans (top left) and Carya, two morphs show complementary temporal separation between male and female flowering (heterodichogamy). Mating types are controlled by two nonhomologous single-locus mechanisms that arose in the common ancestor of each genus, respectively. (Bottom left) Simplified schematic of a putative functional regulatory element at the Juglans locus. (Bottom right) Strong genotypic correlations across the Carya locus indicate a lack of recombination between two colinear haplotypes with similar gene content."
Congrats to @jeffgroh.bsky.social et al on publication of "Ancient structural variants control sex-specific flowering time morphs in walnuts and hickories"
www.science.org/doi/10.1126/...