
CACHE 7 is launched with support from the @gatesfoundation.bsky.social and unpublished data from Damian Young at @bcmhouston.bsky.social, Tim Willson @thesgc.bsky.social and Neelagandan Kamaria InSTEM. Design selective PGK2 inhibitors. We'll test them experimentally.
bit.ly/4lnVYOs
12.08.2025 19:07 — 👍 10 🔁 6 💬 0 📌 0
Check how we are going to transform early-stage hit-finding into a data-driven, computational process driven by open science.
www.nature.com/articles/s41...
23.07.2025 17:30 — 👍 0 🔁 0 💬 0 📌 0
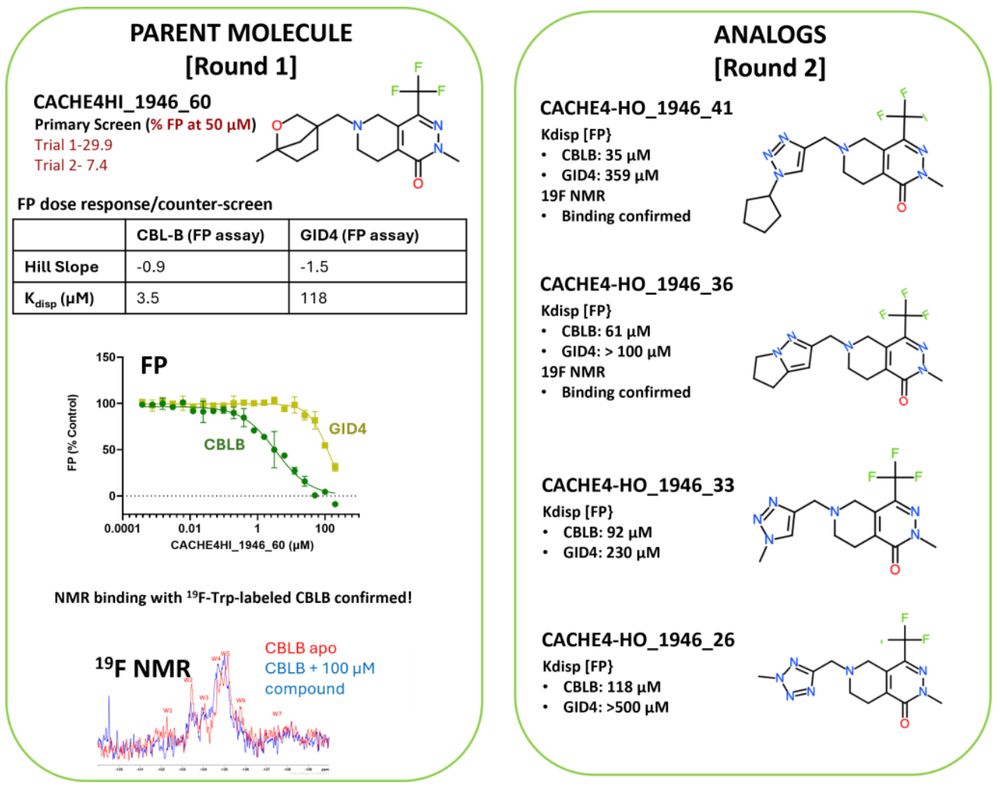
CACHE4 results are out! All previously known CBLCB ligands shared the same scaffold. Congrats to Keunwan Park who successfully designed a chemically novel series, to the experimental team at @thesgc.bsky.social and thanks to @conscience-network.bsky.social for greasing the wheels! bit.ly/4mYNe3r
16.06.2025 16:14 — 👍 8 🔁 5 💬 1 📌 0

First DREAM Target 2035 Drug Discovery Challenge
'First DREAM Target 2035 Drug Discovery Challenge' (Synapse ID: syn65660836) is a project on Synapse. Synapse is a platform for supporting scientific collaborations centered around shared biomedic...
@thesgc.bsky.social is generating large/open screening data and inviting data scientists to train their ML models via DREAM challenges:
1- train your model on DEL data
2- retrospectively predict 138 ASMS true positives
3- predict new hits. We will test them and publish together.
bit.ly/3YXVKoT
05.05.2025 13:58 — 👍 5 🔁 5 💬 0 📌 0

AI drug development’s data problem
The future of drug discovery may be artificial intelligence (AI), but its present is not. AI is in its infancy in the field. To help AI mature, developers need nonproprietary, open, large, high-qualit...
Advancing AI-driven drug discovery begins with open, high-quality data. A great editorial with the authors highlighting how openly accessible datasets have historically accelerated scientific breakthroughs and spotlighting our open data platform: aircheck.ai
More: www.science.org/doi/10.1126/...
17.04.2025 13:41 — 👍 2 🔁 0 💬 0 📌 0

First DREAM Target 2035 Drug Discovery Challenge
'First DREAM Target 2035 Drug Discovery Challenge' (Synapse ID: syn65660836) is a project on Synapse. Synapse is a platform for supporting scientific collaborations centered around shared biomedic...
Advancing ML and drug discovery. @thesgc.bsky.social is now starting to generate protein-ligand data at scale. We’re partnering with CASP, CACHE and DREAM/Sage to run challenges on these data, and help critically assess computational hit-finding methods. Participate!
www.synapse.org/Synapse:syn6...
31.03.2025 20:49 — 👍 4 🔁 1 💬 0 📌 0

In the past two decades, we've distributed over 5,200 plasmids through Addgene, supporting research at 1,084 organizations in 55 countries.
18.03.2025 13:14 — 👍 2 🔁 1 💬 0 📌 0

Episode 5: CACHE – Benchmarking Computational Hit Discovery
Target 2035 Podcast series · Episode
Want to learn more?
🎙️ The Target 2035 podcast breaks down CACHE challenges: open.spotify.com/episode/0bLH...
🎤 The Conscience Symposium on Open Drug Discovery
will feature CACHE participants: conscience.ca/symposium2025
🔗 Learn more about CACHE Challenges: conscience.ca/cache-challe...
17.03.2025 14:44 — 👍 0 🔁 0 💬 0 📌 0
🚀 100 scientists. 31 countries. And we’re just getting started.
#MAINFRAME is uniting global experts to drive AI-powered hit finding. ML models trained on real experimental data. Predictive tools tested in the lab.
🔗 Now is the time to join: aircheck.ai/mainframe
13.03.2025 14:03 — 👍 5 🔁 4 💬 0 📌 1

Nicola Burgess-Brown Webinar Announcement
Join us for the next ProteoCure Webinar on March 20th at 1PM (CET)! Nicola Burgess-Brown from @thesgc.bsky.social will discuss how you can contribute your proteins for screening and obtain protein ligands! Learn more here: proteocure.eu/proteocure-w.... Don't miss out!
10.03.2025 15:46 — 👍 2 🔁 2 💬 0 📌 0
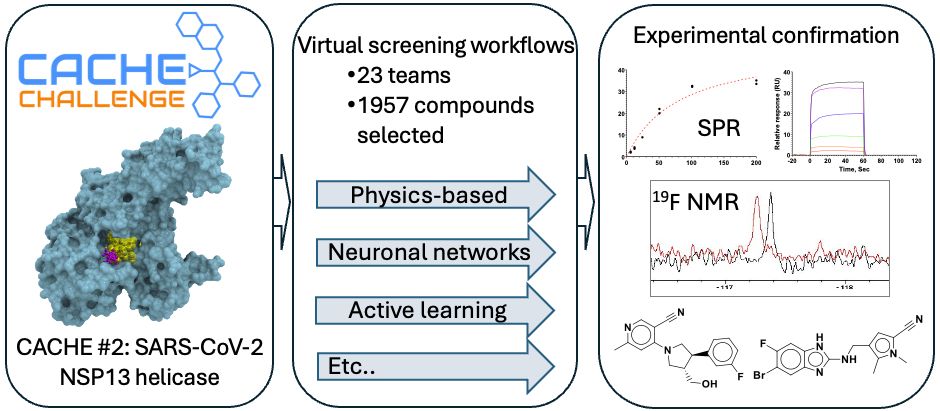
The CACHE #2 preprint is now online: bit.ly/3DyCmHN Active learning and fragment growing delivered confirmed hits. A citizen scientist using the Fold-it gaming interface designed the top compound! Kudos to Sasha and Madhushika @thesgc.bsky.social at the bench. @conscience-network.bsky.social
10.03.2025 21:40 — 👍 7 🔁 7 💬 0 📌 2

The Chemical Probes Portal now covers 601 protein targets, expanding resources for chemical biology & drug discovery! 🧪🔬 New high-quality probes target DCTPP1, MASTL, HCN1, plus E3 ligases & PROTACs for protein degradation. Learn more: www.chemicalprobes.org/news/600-tar... #ChemicalProbes
05.03.2025 09:37 — 👍 1 🔁 1 💬 0 📌 0

Structural Chemistry of Helicase Inhibition
Helicases are essential motor enzymes that couple nucleoside-triphosphate hydrolysis with DNA or RNA strand unwinding. Helicases are integral to replication, transcription, splicing, and translation o...
A new study revealed 4 distinct mechanisms through which small molecules can inhibit helicase activity. Through their analysis, they identified key binding pockets that could be targeted for drug discovery and development for cancer and infectious diseases.
pubs.acs.org/doi/10.1021/...
24.02.2025 17:30 — 👍 1 🔁 0 💬 0 📌 1
Aircheck
Aircheck for Artificial Intelligence-Ready CHEmiCal Knowledge bases
The AIRCHECK Workshop isn't just another conference! It is an exclusive opportunity to learn, experiment, and get ahead of the curve in ML/AI-driven hit discovery.
Register now: event.fourwaves.com/aircheckwork...
19.02.2025 14:47 — 👍 1 🔁 0 💬 0 📌 0
An inspiring story that we are extremely proud to be part of! Kudos to David Drewry and Opher Gileadi who worked along with Paul Workman to identify small molecules that target a historically undruggable transcription factor.
Read their publication: www.nature.com/articles/s41...
18.02.2025 18:39 — 👍 6 🔁 0 💬 0 📌 0
Bridging the Gap: CrossTALK Bootcamp Unites Computational and Experimental Scientists for Drug Discovery | Structural Genomics Consortium
By Sofia Melliou
Can you love computational work and experimental research at the same time? @mattschap.bsky.social and
@rjharding.bsky.social have kicked off the first CrossTALK Bootcamp and they are ready to bring together the next generation of drug discovery experts!
www.thesgc.org/blogs/bridgi...
14.02.2025 15:39 — 👍 0 🔁 0 💬 0 📌 0
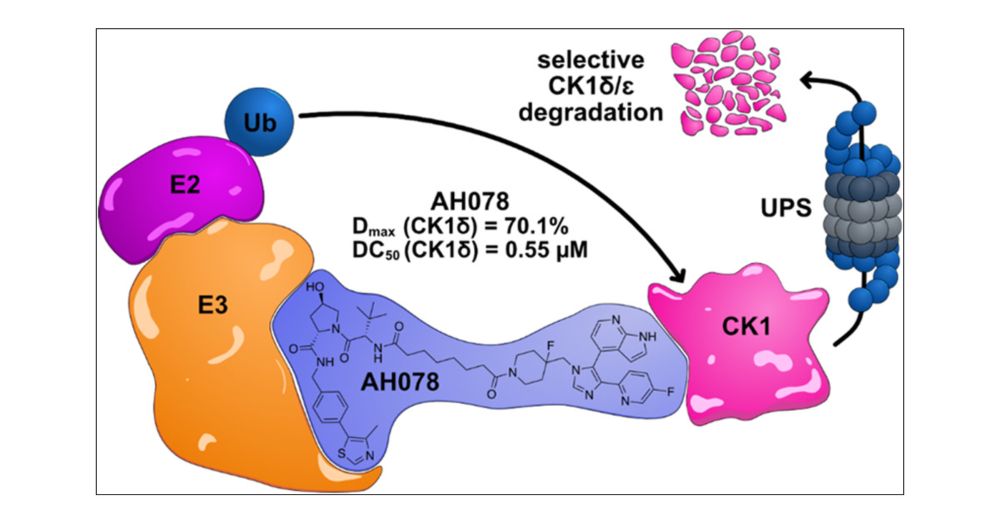
Development and Discovery of a Selective Degrader of Casein Kinases 1 δ/ε
Members of the casein kinase 1 (CK1) family have emerged as key regulators of cellular signaling and as potential drug targets. Functional annotation of the 7 human isoforms would benefit from isoform...
🥳New paper from the lab!🍾 Excited to share our work with the Stefan Knapp lab (Goethe Uni, @thesgc.bsky.social & DKTK), published in J. Med. Chem. @pubs.acs.org. Here, we present the first degraders (PROTACs) targeting CK1 δ/ε—highly selective across the kinome & within the Casein kinase 1 family. 🧵
07.02.2025 13:27 — 👍 5 🔁 2 💬 1 📌 0

We are uniting experts in ML, computational chemistry, and drug discovery to get hands-on experience with AIRCHECK. Co-developed by SGC and @uhntoronto.bsky.social, AIRCHECK is an open data platform designed to enhance predictive models in drug discovery.
event.fourwaves.com/aircheckwork...
05.02.2025 14:45 — 👍 1 🔁 3 💬 0 📌 0

Liver X receptor unlinks intestinal regeneration and tumorigenesis - Nature
Liver X receptor drives epithelial Areg-mediated intestinal regeneration, while preventing tumour growth through adaptive immune responses.
>20 yrs ago, while at GW, now @thesgc.bsky.social UNC scientist Tim Willson and his team invented (and then shared) LXR agonist GW3965. And that GW3965 #chemicalprobe is still having an impact, including in a new study in which we played a small role.
www.nature.com/articles/s41...
30.01.2025 11:15 — 👍 1 🔁 1 💬 0 📌 0


A lovely lunch of Arrowsmith lab and @thesgc.bsky.social friends celebrating Lunar New Year.
31.01.2025 02:18 — 👍 4 🔁 1 💬 0 📌 0
Brain Canada is a national non-profit organization that enables and supports excellent, innovative, paradigm-changing brain research in Canada. #BrainCanada
Welcome to #UofT! Canada’s top university, and a catalyst for discovery, innovation, and progress. #UofTDefyGravity
🔗 utoronto.ca
Transforming genomics research into solutions for Canada | Transformer la recherche en génomique en solutions pour le Canada genomecanada.ca | genomecanada.ca/fr
The Leslie Dan Faculty of Pharmacy at the University of Toronto is one of North America's leading pharmacy educators and a hub for innovation and discovery.
Welcome to the Department of Chemistry at the University of Toronto. Follow for student info, faculty awards, alumni news & the latest events, seminars & colloquia.
Research news from Canada's largest research hospital.
🔗 uhnresearch.ca
Expert recommendations and info about #ChemicalProbes, particularly small molecule inhibitors, and their use in biomedical research and #DrugDiscovery.
Compte officiel du Neuro (Institut-Hôpital neurologique de Montréal)
Official account of The Neuro (Montreal Neurological Institute-Hospital)
The ICR is making the discoveries that defeat cancer.
We're a top UK academic research centre, a postgrad college, and a charity.
📍 London (Chelsea and Sutton)
https://www.icr.ac.uk/
Prof @ U. Toronto - PI @ SGC - CACHE challenges - Computational chemistry- Structural bioinformatics - Biophysics
chemical biology enthusiast & strategist; I make impossible things possible - all before breakfast!
discovery chemistry nerd @Merckgroup; oncology, immunology, targeted protein degradation, ADCs; soccer, politics, tapas can slip into the feed; my own views
Scientist. Frog hunter. Founder and Chief Executive of Structural Genomics Consortium.
Bringing health and economic benefits to the people of Ontario through world-leading cancer research. Views expressed are those of OICR.
Read the latest news from OICR at: https://oicr.on.ca/news/
Canada's Hospital:
Princess Margaret, Toronto General, Toronto Western, Toronto Rehab, West Park Healthcare & Michener.
🔗 uhn.ca
Advancing drug discovery through #AI, #OpenScience, and #Collaboration between academia, industry, and non-profits to create accessible treatments for all. More about us at conscience.ca
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app



























