Still time to submit an abstract for our free online workshop as part of COEXIST🧪🦣🏺
07.01.2026 07:22 — 👍 10 🔁 11 💬 0 📌 0

🚨ONLINE WORKSHOP🚨
We will be hosting a free online workshop “Maximising Data from Palaeolithic Bone" as part of the the COEXIST project on 10th March 2026. Please submit abstract by 23rd January 2026. Further information in the flyer. See you there!
15.12.2025 09:33 — 👍 19 🔁 21 💬 0 📌 1
AT ICAZ AGPM (Copenhagen) @wagneranna.bsky.social will give a talk contrasting her current #ZooMS results from Germany and Romania, while @geoffreymsmith.bsky.social will present his #ZooMS results from the British Middle Pleistocene site of Victoria Cave. @icaz-news.bsky.social
14.08.2025 13:36 — 👍 1 🔁 1 💬 0 📌 0
@eshesociety.bsky.social (Paris) both COEXIST PhD students will be presenting posters on faunal assemblages in Central and Southeast Europe, @frankietait.bsky.social on refining chronologies, and @wagneranna.bsky.social on rethinking subsistence integrating #Zooarchaeology and #ZooMS.🧪🦣🏺
14.08.2025 13:36 — 👍 0 🔁 0 💬 1 📌 0
In our EAA session, we will give 3 talks including an overview of our project aims and progress, and two case studies presenting our #ZooMS data from Crvena Stijena (Montenegro) and Cioarei Borosteni (Romania).
14.08.2025 13:36 — 👍 2 🔁 0 💬 1 📌 0
@archaeologyeaa.bsky.social (online), we are organising an entire session on Neanderthal-Homo Sapiens subsistence differences 60,000 to 40,000 years ago in Central and Southeast Europe
14.08.2025 13:36 — 👍 0 🔁 0 💬 1 📌 0
At @isbarchaeology.bsky.social (Turin), @frankietait.bsky.social will be presenting our ZooMS identifications and new C14 dates from Middle Palaeolithic layers in the cave site of Cioarei Borosteni (Romania) @isba11.bsky.social
14.08.2025 13:36 — 👍 1 🔁 0 💬 1 📌 0
We are approaching conference season! We are currently preparing 9 presentations for 4 conferences, and look forward to sharing some of our first COEXIST project results, and more! #archsci #Palaeoproteomics #ZooMS #Zooarchaeology 🧪🦣
14.08.2025 13:36 — 👍 6 🔁 1 💬 1 📌 0
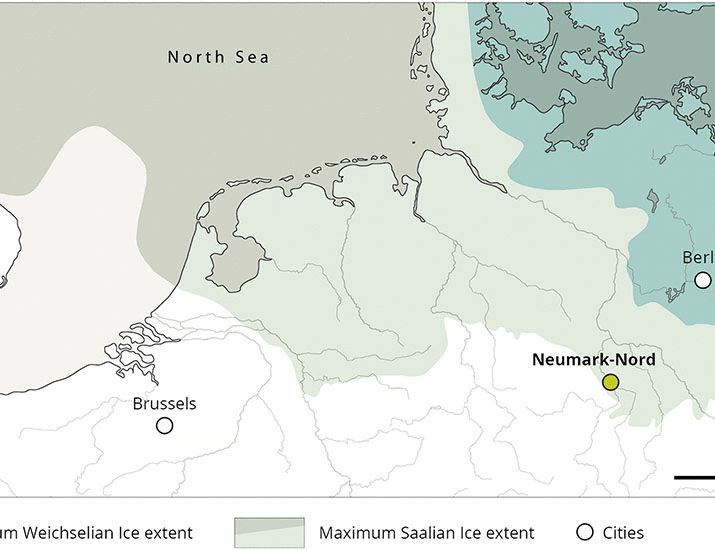
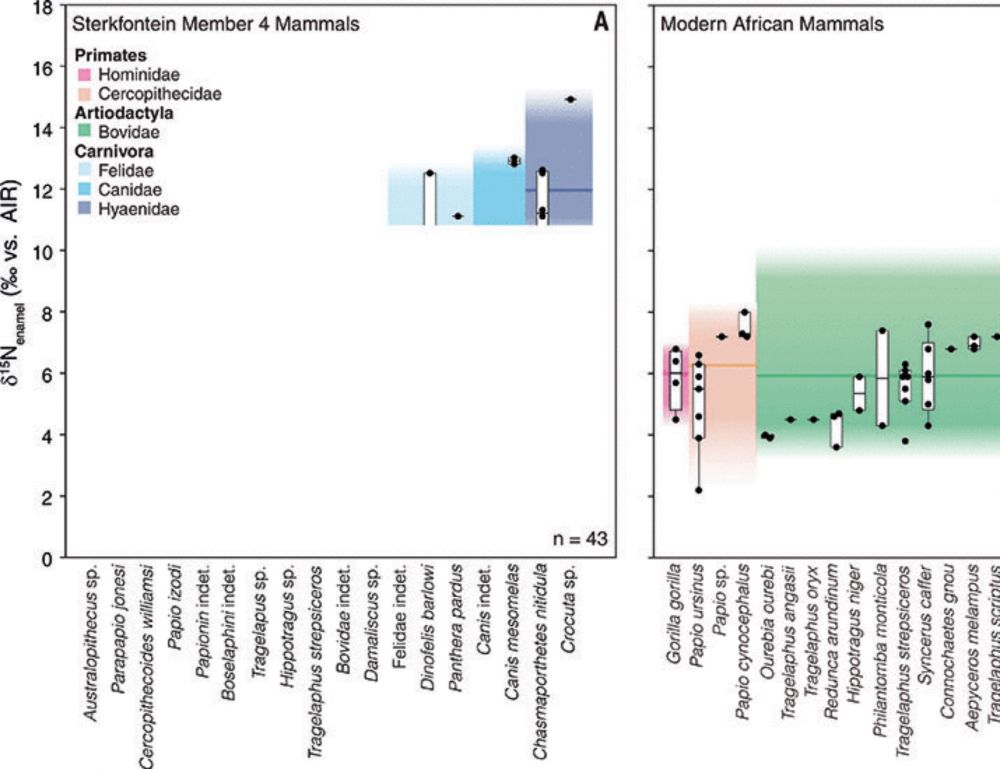
Large-scale processing of within-bone nutrients by Neanderthals, 125,000 years ago
Neanderthals intensively processed a minimum of 172 large mammals for grease and marrow fat, 125,000 years ago.
New paper in #ScienceAdvances led by @paleomonrepos.bsky.social and @unileiden.bsky.social on #Neanderthal adaptation 125ka years ago at Neumark-Nord 2, Germany. Our study shows large-scale grease rendering thousands of years earlier than previously thought: www.science.org/doi/10.1126/... 🦣🧪🏺
03.07.2025 05:30 — 👍 27 🔁 8 💬 1 📌 0

Spotting her first MALDI plate! Thanks to @archaeoprotein.bsky.social for offering this wonderful training opportunity to @sarahbarakat.bsky.social and helping us further develop our ZooMS capacity in Aberdeen @uoa-archaeology.bsky.social - and for running our first PALaEoScot samples with us!
02.07.2025 12:23 — 👍 20 🔁 5 💬 0 📌 1
This interactive 1.5 hour webinar allows students and researchers from around the world to ask questions from our panel of #ZooMS and #Zooarchaeology experts: @geoffreymsmith.bsky.social, Camilla Speller, @abigaildesmond.bsky.social, Emmanuel Discamps, @mcodlin.bsky.social and Naihui Wang.
15.05.2025 07:55 — 👍 5 🔁 2 💬 1 📌 0

We are hosting a free online ‘Ask Us Anything’ event as part of the Integrating ZooMS and Zooarchaeology workshop series. For more info and to submit questions: docs.google.com/forms/d/e/1F...
📅 Date: June 30, 2025
🕒 Time: 15:00 CET
📍 Location: Online (link: univ-tlse2.zoom.us/j/9775805212...)
15.05.2025 07:45 — 👍 19 🔁 16 💬 2 📌 9

ICAZ AGPM 2025
Exploring the Past, Informing the Future: �Two Decades of Interdisciplinary Approaches in Archaeozoology
The next ICAZ-AGPM meeting (Archaeozoology, Genetics, Proteomics and Morphometrics) will take place 14-17 October 2025 in Copenhagen, Denmark 🐘🐐🦈🦜
Abstract are due 25 April!
More info: sites.google.com/palaeome.org...
18.03.2025 13:24 — 👍 9 🔁 8 💬 0 📌 5

Simplified workflow for the collagen extraction procedures and mass spectrometry techniques (MALDI-TOF and MALDI-FTICR) applied to a sample of indeterminate bone fragments from Le Piage, France. This figure illustrates the three collagen extraction protocols (AmBic, TFA and HCl) and the two mass spectrometry approaches (MALDI-TOF and MALDI-FTICR) used in the analysis. The lower left and right panels show Peptide Mass Fingerprinting (PMF) of peptide P1 (ɑ1 508, m/z 1105) for a single sample (PI-134-ICR) analysed with both mass spectrometers. The isotopic distributions are markedly different in these panels due to the separation by MALDI-FTICR of the two components of the first isotope. The spectrum shows a 13C and deamidated peptide peaks separated by 0.019 m/z.
🧵 Thread: Comparing mass analyzers for ZooMS on poorly preserved collagen 1/5
Raymond et al. @palaeocdf.bsky.social @archaeoprotein.bsky.social test MALDI-TOF vs MALDI-FTICR for ZooMS analysis on Palaeolithic bone fragments (37-34 ka BP).
Paper: doi.org/10.1002/rcm....
#ZooMS
16.03.2025 09:02 — 👍 10 🔁 6 💬 1 📌 0
The #palaeoproteomics #ZooMS starter pack has been updated to include, amongst others
@archaeoprotein.bsky.social and @drsambrown.bsky.social
06.02.2025 21:12 — 👍 8 🔁 5 💬 0 📌 0
The @eaapam.bsky.social deadline has been extended until 10th February. Still time to submit to our session on #Zooarchaeology #ZooMS and subsistence in central and SE Europe. 🦣🧪🏺
06.02.2025 11:56 — 👍 9 🔁 8 💬 0 📌 0

Mystery woman’s DNA reveals close family ties between Europe’s earliest people
Pair of studies shines light on how modern humans and Neanderthals settled the continent together
New findings pin down when the Zlatý kůň woman lived to about 45,000 years ago and shed light on the remarkably mobile lifestyle of the earliest groups of modern humans to enter Europe. scim.ag/3C0ZVbp
06.01.2025 18:05 — 👍 294 🔁 46 💬 9 📌 5
4/5 Key challenge: We need "mediators" - scholars who understand both molecular science AND humanities. Current education systems need updating to bridge this gap. The future demands experts who can speak both languages! 🔬📚
17.01.2025 20:40 — 👍 22 🔁 2 💬 3 📌 1
For those of you troubled by the JHE resignations, PaleoAnthropology provides a free #openaccess alternative, one of the reasons we published our #ZooMS Special Issue there.
13.01.2025 14:53 — 👍 2 🔁 0 💬 0 📌 0

Journal editors’ mass resignation marks ‘sad day for paleoanthropology’
Exodus from the Journal of Human Evolution leaves a flagship journal in crisis
Science reporting on the mass resignation of the editorial board of the Journal of Human Evolution, highlighting some of the problems with commercial scientific publishing.
OA PaleoAnthropology journal (www.paleoanthropology.org) highlighted among JHE's successors
#openaccess #humanevolution
10.01.2025 12:22 — 👍 53 🔁 28 💬 2 📌 2
Follow us here to stay up to date with the latest news from our new Archaeological Proteomics Lab at the University of Reading. We will keep you updated with news from our various projects and our current publications
#palaeoproteomics #zooarchaeology #ZooMS 🦣🧪🏺
09.01.2025 16:07 — 👍 13 🔁 6 💬 0 📌 1
(Geo)Archaeologist into 'dirt' and human behavior-related things.
PI of ERC-StG MATRIX https://matrix.icarehb.com/
A centre of excellence in bioarchaeology, specialising in the analysis of human, animal and plant remains and molecular techniques - proteins, lipids, DNA and stable isotopes. Department of Archaeology, University of York
Fyssen Foundation post-doctoral fellow | University of Bordeaux | FR |
PACEA - BordeauxProteome - CBMN
#Palaeoproteomics #ZooMS #archsci #bioarch
Ancient proteins and Palaeolithic Zooarcheology
Scientist living in Vienna. Two^H^H^H Three loud kids, population genetics, sediment DNA. New Prof. @ Uni Vienna / Group leader @ MPI EVA. ERC CoG UNEARTH.
Palaeoproteomics Researcher at CENIEH
🧪👩🔬🦴🦷🌏
Building a brand new #palaeoproteomics laboratory here in Burgos, Spain. Exciting new things on the horizon!
#archaeology #evolution #palaeolithic #ZooMS #proteins
Proteomics | mass spec | scientific illustration | Magic: the Gathering
🔬 Archaeologist interested in how stone tools were used 🖐️Researcher at: @paleomonrepos.bsky.social @leizarchaeology.bsky.social
@tracer-leiza.bsky.social
@icarehb.bsky.social (www.joaomarreiros.com)
This it the official Bluesky account for the archaeology department at the University of Aberdeen. Follow us to see what we are up to (or digging down into)
Associate Professor in Osteology at the University of Bergen 🇳🇴 - Zooarchaeology, Historical Ecology, and Palaeoproteomics - Cetaceans 🐳 and South American Camelids 🦙 - 🇳🇱 (he/him)
Doctoral Researcher in Archaeological Sciences at @univie.ac.at -
Department of Evolutionary Anthropology. Exploring Late Neanderthal sites in the Palaeolithic - Radiocarbon dating, Paleoproteomics, Stable Isotopes. Yorkshire lass in Vienna.
Palaeoproteomics
Currently in Denmark
She/Her
Postdoc
Archaeologist. Mom. Anthropology faculty at University of Texas at El Paso. Zooarchaeology, Paleoecology, Women in science. She/Her/Hers
PhD Candidate @UCSB | Zooarchaeology | Isotopes | Mississippian Archaeology | Andes | California |
I 💙 llamas
https://snoe00.wixsite.com/sarah-j-noe
Interested in ancient proteins & Human Evolution | @Welkergroup.bsky.social | Globe Institute, University of Copenhagen | 🦷🦴💀
Funded by the European Research Council (No. 949330). In collaboration with the FECYT– Ministerio de Ciencia, Innovación y Universidades
Lab specialized in the study of macro- and microscopic wear traces and residues on prehistoric stone artefacts. University of Liège (BE).
Visit us @Traceolab.be
Prehistoric archaeologist and hunter-gatherer of interestingness 🏹🌾 | Sharing my love of all things ancient 🏺🏛️ | PhD student at Cambridge studying the Neanderthals of Shanidar Cave, Iraqi Kurdistan
https://linktr.ee/oz_of_the_ancients
Official bluesky account of the European Society for the study of Human Evolution. Email us communication@eshe-conference.eu
www.eshe.eu
Official Bluesky account for PaleoAnthropology
Published jointly by the Paleoanthropology Society and the European Society for the Study of Human Evolution (ESHE)
paleoanthropology.org