How do dynamics of signalling molecules skew cell fate decisions? With this review we try to move our first steps in the signalling dynamics field, together with the expert lead of @samuzambrano.bsky.social! Kudos to Fulvio, Sara and Erika for their excellent work!
23.10.2025 18:02 — 👍 2 🔁 1 💬 0 📌 0
Slightly diminish a band:
They might be quite tall.
26.09.2025 15:55 — 👍 1 🔁 0 💬 0 📌 0
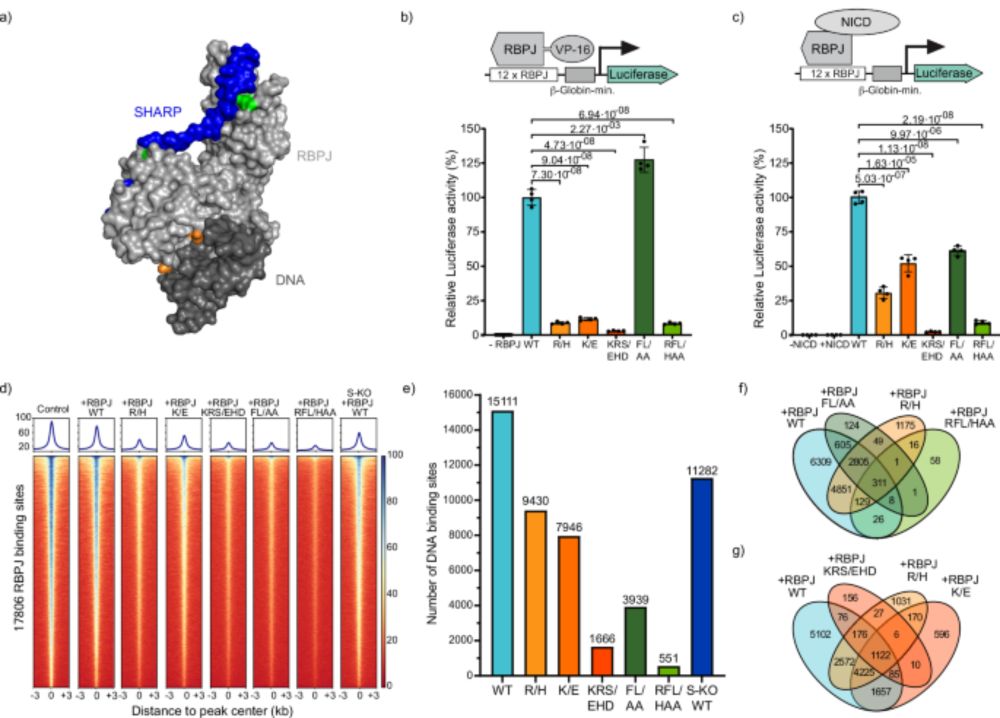
Minflux goes to chromatin! Very interesting preprint from @andersshansen.bsky.social lab, featuring co-first author @matteomazzocca.bsky.social, that keeps on making me very proud.
15.05.2025 07:41 — 👍 4 🔁 0 💬 0 📌 0
That's so great, Davide! Congratulations!
07.03.2025 08:10 — 👍 1 🔁 0 💬 0 📌 0
This looks sooo great! Congratulations @mirlab.bsky.social !!
12.02.2025 13:30 — 👍 6 🔁 1 💬 0 📌 0
Great to see you here, Tim!
07.02.2025 19:58 — 👍 0 🔁 0 💬 1 📌 0
Fantastic Work Christof!
07.02.2025 19:55 — 👍 0 🔁 0 💬 0 📌 0
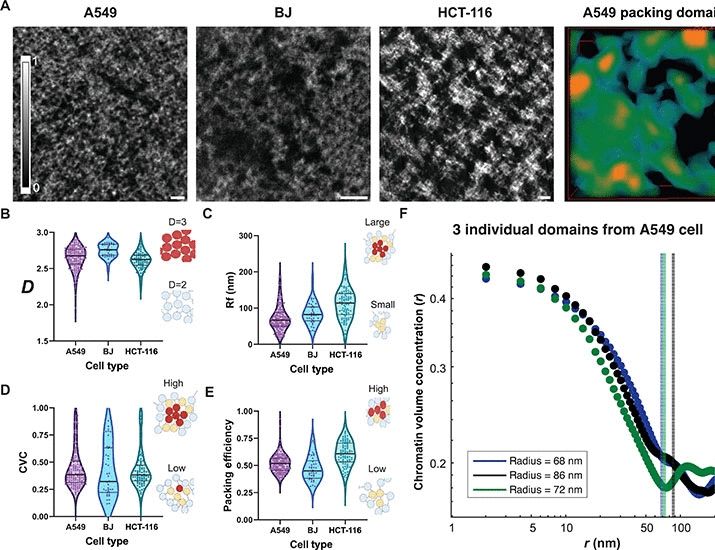
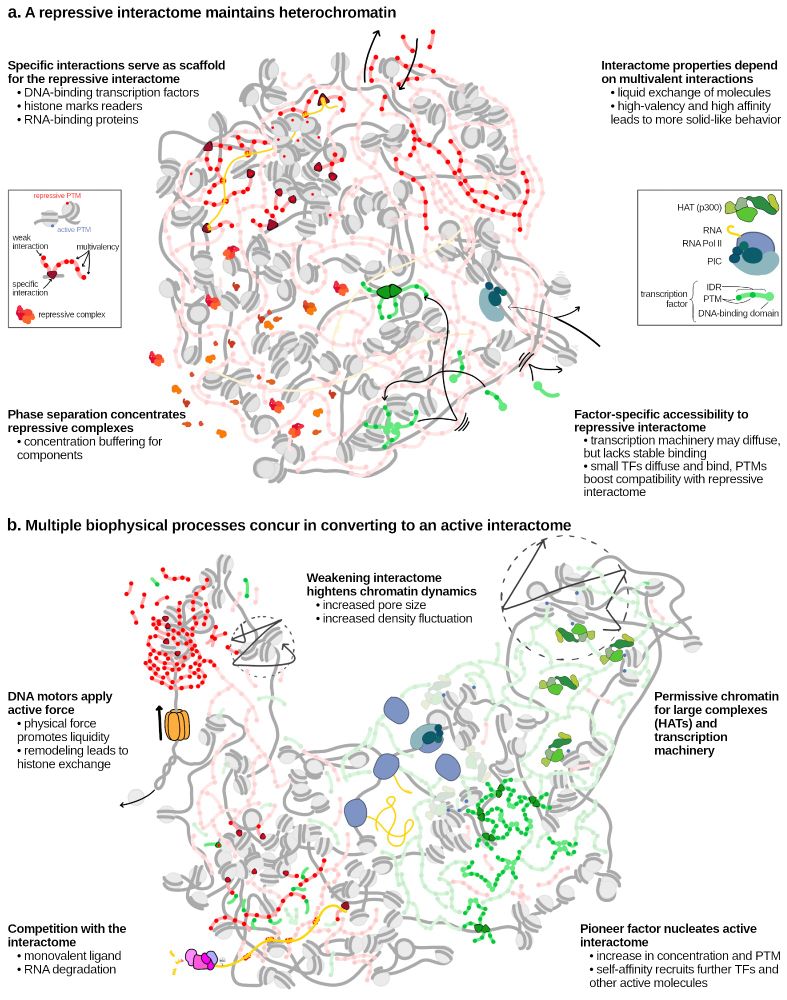
What is the multiscale structure of chromatin condensates? How does it shape thermodynamic and material properties?
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)
23.01.2025 09:32 — 👍 118 🔁 30 💬 7 📌 4
Thank you Laura, you are right! We should have cited that paper!
13.01.2025 16:05 — 👍 1 🔁 0 💬 0 📌 0
Real-time visualization of reconstituted transcription reveals RNA polymerase II activation mechanisms at single promoters https://www.biorxiv.org/content/10.1101/2025.01.06.631569v1
07.01.2025 06:46 — 👍 5 🔁 1 💬 0 📌 1
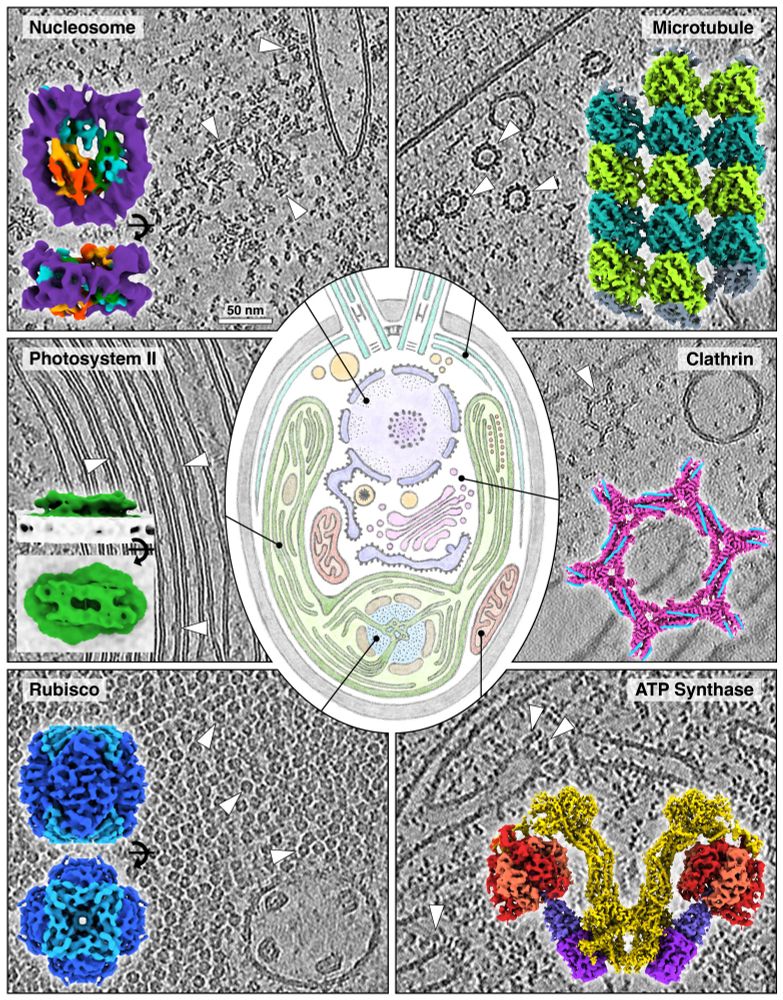
Towards community-driven visual proteomics! Excited to finally share this large-scale curated & annotated dataset of 1829 high-quality #cryoET tomograms of the little green alga that just keeps giving— Chlamydomonas! 🧪🧶🧬🌾🌊🌍
Preprint📜: www.biorxiv.org/content/10.1...
A short thread🧵👇
06.01.2025 11:18 — 👍 337 🔁 128 💬 7 📌 14
Tasty, tasty phase separation. A much needed rigorous study!
04.01.2025 19:45 — 👍 2 🔁 0 💬 0 📌 0
Thank you Kazuhiro.Your work was a great inspiration for this! We hope to have reported it accurately.
23.12.2024 14:14 — 👍 1 🔁 0 💬 1 📌 0
Thanks Mark, all merit goes to Tom on this! He went through many iterations before having something that was somehow 'accurate' and yet readable.
23.12.2024 14:10 — 👍 1 🔁 0 💬 0 📌 0
Thank you Lennart!
20.12.2024 21:57 — 👍 0 🔁 0 💬 0 📌 0
We often speak about chromatin as being accessible or inaccessible, but what does it mean? We wrote a short review on this, 🔬 focused:
sciencedirect.com/science/arti...
A big thank you to Tom Fillot for his efforts on this and to
@hansen_lab
@marcelonollmann
for their help as editors.
20.12.2024 15:46 — 👍 136 🔁 55 💬 4 📌 3
Thank you very much Carmelo! Happy to have joined this lively community!
20.12.2024 15:40 — 👍 1 🔁 0 💬 1 📌 0
Official account of the 2025 retreat of the Experimental Oncology and Genetics & Cell Biology divisions @SanRaffaeleMI
senior scientist at Janelia | single particle tracking & #microscopy 🔬 | PhD on single-molecule enzymatic dynamics | my path: 🇩🇪→🇺🇸→🇸🇪→🇺🇸 | https://scholar.google.com/citations?user=UzGfR3MAAAAJ&hl=en
Ph.D student | Prof. Koby Levy group | Weizmann Institute of Science |
Molecular Dynamics of Biomolecules 🧬
Postdoc @andersshansen.bsky.social Lab @ MIT | PhD @davidemazza.bsky.social Lab @ Vita-Salute San Raffaele University | 3D genome, TFs, gene regulation
Group Leader at Italian Institute of Technology.
RNA and Chromatin lover
Coordinator of λ-lab @ Center for Omics Sciences, Milan | Assistant professor of bioinformatics @unisr.bsky.social
Giornalista. Vicedirettore del Post.
Just another LLM. Tweets do not necessarily reflect the views of people in my lab or even my own views last week. http://rajlab.seas.upenn.edu https://rajlaboratory.blogspot.com
Dedicated to drug discovery, enthralled by science. 5 kids. She. SAB Recursion. Founder SyzOnc. Lab at Broad Institute. Opinions my own.
Scientific instrument developer, focused on #lightsheet and other #microscopy 🔬 at ASI
Scientist interested in genes in living cells. Associate Professor at Colorado State University.
Proprio lui. No un altro.
💻 🧬 3D genome & RNA lovers Group Leader at Center for Human Technologies, Italian Institute of Technology | CDA @ArmeniseHarvard
Physics at Ulm University | live-cell/organism single-molecule microscopy | super-resolution microscopy | gene regulation | chromatin architecture | embryo development
CNRS Senior Researcher - Group Leader at Laboratory of Biology and Modeling of the Cell, Ecole Normale Supérieure de Lyon.
Physical biology of chromatin: modeling the spatio-temporal dynamics of eukaryotic genomes.
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
NeuroCyto lab 🤹🏻 Institute of NeuroPhysiopathology at CNRS & Aix-Marseille Université 🧠 Neuronal cell biology 🦠 Super-resolution microscopy 🔬 Come for science 👨🏻🔬 Stay for some cooking, sneakers and bike adventures 🥘 👟🚴
Postdoc in the Medema-Vader lab working on extrachromosomal DNA 🧬 | Prinses Máxima Center, Utrecht
🇪🇺 MSCA Fellow
🔬Former PhD in the Rippe lab | DKFZ & Heidelberg University