
APSPM 2026: Structural Phylogenetics Meeting
A pivotal SMBE regional meeting in Brisbane on the interface of protein structure, function, and evolution.
Meet the plenary speakers for #APSPM2026.
Joanna Masel @joannamasel.bsky.social
Nicholas Matzke @nicholasmatzke.bsky.social
Join us to discuss how protein structure is changing evolutionary theory, phylogenetics and molecular evolution.
biosig.lab.uq.edu.au/strphy26/spe...
14.11.2025 02:23 — 👍 3 🔁 2 💬 0 📌 0
2026 Astrobiology Science Conference
Abstract submission deadline for #AbSciCon2026 is Jan 14. I'll be co-organizing a session on "The Last Universal Common Ancestor and the Origins of Translation" with @lorendw.bsky.social and @seekingluca.bsky.social, with lots of other cool related topics at agu.confex.com/agu/abscicon.... Join us!
13.11.2025 18:26 — 👍 3 🔁 1 💬 0 📌 0
An empirical approach to evaluating the prevalence of long-lived balancing selection in humans--and important limitations. Work by @hannahmm.bsky.social
11.11.2025 19:14 — 👍 62 🔁 35 💬 0 📌 0
🧪
Hard to believe that this is still something we have to argue for in 2025. The science is settled.
28.10.2025 21:08 — 👍 25 🔁 10 💬 1 📌 0

APSPM 2026: Structural Phylogenetics Meeting
A pivotal SMBE regional meeting in Brisbane on the interface of protein structure, function, and evolution.
We are excited to announce APSPM 2026 - the SMBE Australasian Protein Structural Phylogenetics Meeting!
🗓️ 15-18 Feb. 2026
🌏 Brisbane, Australia
🔗 biosig.lab.uq.edu.au/strphy26/
📄 Abstract submission: 28.11.2025
✈️ Travel grant application: 28.11.2025
✏️ Registration: 15.01.2026
#StrPhy26
22.10.2025 10:20 — 👍 8 🔁 8 💬 1 📌 0
The new #NSF #GRFP application now excludes 1000’s of students, including those who were told they could apply this year, or who planned to apply next year.
We have a new specific petition urging #NSFGRFP undo this harmful action. If you are impacted sign here: laurenkuehne.github.io/grfpChanges/
29.09.2025 17:47 — 👍 21 🔁 33 💬 2 📌 8

Universal adaptive strategy theory - Wikipedia
Tilman posited that apparently low-density environments (where Grime let ruderals succeed) had larger territories, with resources still exploited down to the lowest tolerated level. We support Grime’s view, by highlighting that resource levels need not be so low en.wikipedia.org/wiki/Univers... 9/9
16.09.2025 17:37 — 👍 4 🔁 0 💬 0 📌 0
Redirecting
In previous work, we tackled different ways of competing over territories, i.e. durable resources doi.org/10.1016/j.tp.... Our current work addresses consumable resources. Both can be seen as formalizations of Grime’s ruderals vs competitors vs colonizers. 8/9
16.09.2025 17:36 — 👍 2 🔁 0 💬 1 📌 0

Series of phase diagrams showing when coexistence vs one type winning occurs, and via which mechanism. Each phase diagram has the hawkishness of the two types as its axes, based on the rate at which they give up during contests. Different panels vary in their search and/or handing rates, changing both exploitation and interference.
The same difference in exploitative ability yields more coexistence when created by search speed differences than via handling times. 7/9
16.09.2025 17:36 — 👍 2 🔁 0 💬 1 📌 0
Faster search yields more opportunities to interfere, and faster handling yields fewer opportunities to be interfered with. This mechanistic connection between exploitative and interference abilities is core to our results, and how they differ from past work. 6/9
16.09.2025 17:36 — 👍 1 🔁 0 💬 1 📌 0

Table showing the conditions for six different outcomes: the R* rule, Hawk victory, Dove victory, Hawk-Dove coexistence, the Dominance-discovery trade-off, or the Dove-discovery trade-off. Terms for the strength of Exploitative differences vs Interference differences enable us to distinguish among the scenarios.
Other parameters yield the “dominance-discovery” trade-off described in ants, where the Dove loses at contests but is better at finding new resources. We also find a new “Dove-discovery” trade-off, where Hawks search better, but the opportunity costs from contests is too high. 5/9
16.09.2025 17:35 — 👍 3 🔁 0 💬 1 📌 0
Some parameter choices yield something like the Hawk-Dove game, with a trade-off between the benefits of interfering with the other type and the costs of interfering with one’s own type. The Hawk’s cost need not be direct – an opportunity cost from time spent fighting is enough. 4/9
16.09.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0
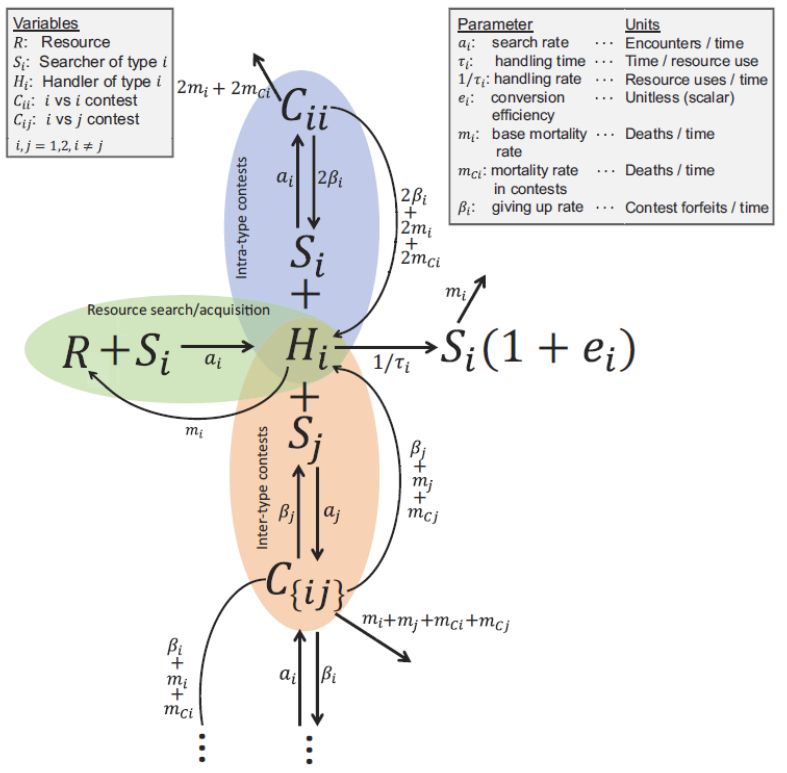
Ball and stick diagram showing mass action kinetics by which Searchers find Resources and coming Handlers, Searchers find Handlers to begin a Contest, Contests end when one side dies or gives up, Handling ends with consumption, or Handlers or Searchers die.
We develop a mechanistic model in which consuming a resource takes time, and Handlers can be interrupted by Searchers who find them rather than free Resource. This initiates a Contest, distracting the consumers, thus allowing the resource to grow to higher levels. 3/9
16.09.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0
R* rule (ecology) - Wikipedia
R* theory assumes no direct interactions at the same trophic level, and that resources are consumed down to the level at which only one species can persist. en.wikipedia.org/wiki/R*_rule... 2/9
16.09.2025 17:33 — 👍 1 🔁 0 💬 1 📌 0
The House discretionary budget bill is now out for FY26, including a proposed *slight boost* in NIH funding & many other components that will take time to unpack.
Bill text:
appropriations.house.gov/sites/evo-su...
A few key points including IDC analysis in a provisional 🧵
02.09.2025 00:43 — 👍 38 🔁 34 💬 1 📌 5

Trump Deals A New Immigration Blow To International Students
Trump officials have proposed a new rule limiting international students to fixed periods of entry, making a U.S. education more precarious.
Trump admin planning to change student visas from lasting for duration of academic program to fixed 4-yr term, and then much harder to renew
Could destroy US ability to attract global talent, particularly those seeking advanced degrees in STEM. The median time to complete a PhD is 5.7 yrs per NSF.
29.08.2025 10:52 — 👍 1498 🔁 708 💬 97 📌 152

Viruses are a dominant driver of protein adaptation in mammals
Viruses drive adaptation at the scale of the whole proteome and not only in antiviral proteins in mammalian hosts.
I think the strongest evidence for pervasive adaptive substitutions comes from @denard.bsky.social eg elifesciences.org/articles/12469 and subsequent work. The reason is pathogens, although other arms races might contribute.
05.08.2025 02:59 — 👍 0 🔁 0 💬 0 📌 0
I think there is a drift barrier in all cases. But allele frequencies trajectories are different, affecting the site frequency spectrum. We found that the Ne reduction stems from interactions between beneficial and deleterious alleles: bsky.app/profile/joan...
05.08.2025 02:55 — 👍 0 🔁 0 💬 0 📌 0
Whoops, I just realized what you were pointing to. I was quoting @nicolasgaltier.bsky.social that time. It's really about the effects of sites that the focal allele is in LD with: either fewer of them and linked, or potentially weakly the whole genome.
05.08.2025 02:49 — 👍 0 🔁 0 💬 1 📌 0
Also, I think what I wrote about linked selection in that thread was a direct quote from the authors.
05.08.2025 02:47 — 👍 0 🔁 0 💬 1 📌 0
You are asking why in a thread I wrote 2 years ago, I wasn't yet aware of all the results that came from our 1-year-old paper!!! I think you can guess the reason...
05.08.2025 02:46 — 👍 0 🔁 0 💬 1 📌 0
Redirecting
The quality of genetic background has temporal autocorrelation. Neutral theory cannot capture the resulting colored noise. www.cell.com/current-biol...
05.08.2025 02:45 — 👍 0 🔁 0 💬 0 📌 0
The theory of background selection from linked sites is very different to that from unlinked sites. Charlesworth actually started with the latter, although most current work is the former.
05.08.2025 02:41 — 👍 0 🔁 0 💬 1 📌 0
The paper finds var(#del mut per individual) < mean. This implies non-independent evolution. Think of selection as taking out the worst quadrant given two arbitrary axes - you'll get a negative correlation after, ie LD among unlinked sites.
05.08.2025 02:40 — 👍 0 🔁 0 💬 1 📌 0
Evolutionary Biologist | Protein Structure Enthusiast | Quantum Programmer | Wannabe Polymath | Science Comm | Life Sciences Programmer | Originally from 🇵🇰
Research fellow @aucklanduni.bsky.social
Structural phylogenetics #strphylo protein evolution, plant systematics
Structural Phylogenetics • methods, datasets, tools, events • concise updates for researchers and practitioners. Managed by @proteinmechanic.bsky.social and @cpuentelelievre.bsky.social
PhD candidate @anja1.bsky.social group @NIOZ and @UvA 🇳🇱. Interested in molecular evolution, Archaea and cell biology. https://w-huang.com
PhD candidate @ Biological Sciences, Columbia
Population genetics & evolutionary genomics
Genome evolution, NC State University
http://conantlab.org
Early career PI and mother
Omidyar Postdoctoral Fellow at the Santa Fe Institute, interested in major evolutionary transitions, cooperation, conformity, and AI. Canadian 🇨🇦 she/her
Evolutionary systems/cell biologist. EMBO and SNSF Postdoctoral Fellow with Dmitri Petrov and Dan Jarosz at Stanford. PhD with Andreas Wagner at the University of Zurich. Studying how molecular and cellular systems shape, and are shaped by, evolution.
Statistical geneticist. Associate Prof at Dana-Farber / Harvard Medical School.
www.gusevlab.org
Evolutionary Biology @ Emory
https://biology.emory.edu/people/bios/faculty/bazykin-georgii.html
Postdoc, UniWien DOME/CMESS
Watching individual bacteria grow and starve
he/him
Postdoc @uwgenome 🐶
PhD Genetics @uarizona 🐱
BA Chemistry @brynmawrcollege 🦉
Population Genomics, Machine Learning, Cancer Evolution 🧬💻
📍Seattle, WA & Vancouver, BC
https://www.linkedin.com/in/linhnhtran/
Professor of Theoretical Biology at the University of Würzburg
Assistant Professor, interested in:
Theory of evolution, Plant Science, mathematical biology, evolution of innovation.
Research Scientist at Carnegie Science
Bioinformatics | Bacterial Natural Products | Host-Microbe interactions












