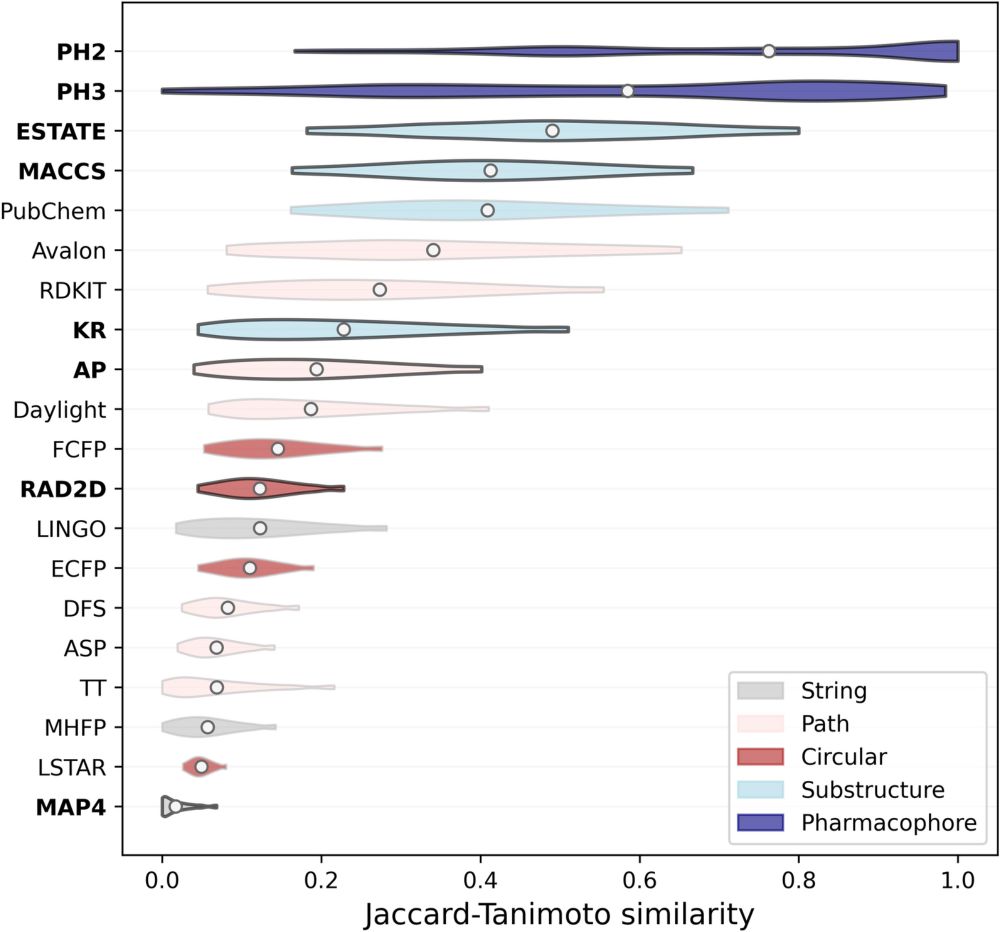
Interesting talk by @fragrisoni.bsky.social at #ChemBioPhys2025. She talked about her group's exciting research into exploring the vast chemical space for #DrugDiscovery using #ArtificialIntelligence in low-data scenarios. www.nature.com/articles/s43...
#ChemBio #ChemSky
23.10.2025 15:47 —
👍 13
🔁 4
💬 0
📌 0
In addition, the workshop will host the annual meeting of the ELLIS Program on Machine Learning for Molecule Discovery, fostering interdisciplinary dialogue and shaping the next phase of molecular AI research.
Organizers:
🔸 Nadine Schneider
🔸 @fragrisoni.bsky.social
🔸 José Miguel Hernández Lobato
10.10.2025 08:42 —
👍 1
🔁 1
💬 0
📌 0

Dream Reactions Symposium – Wrap-Up Part I ✨
Our speakers are truly passionate about driving towards a positive change! 🌍
Special congratulations to @fragrisoni.bsky.social, the very first recipient of the Green Dream Reactions Award!! 🏆👏
#DreamReactions#Sustainability
22.08.2025 08:23 —
👍 16
🔁 4
💬 1
📌 0

It is a great pleasure to share that our extensive review on "The Chemistry and Biology of the Tetrodotoxin Natural Product Family" has been accepted for publication at Angewandte Chemie: doi.org/10.1002/anie....
18.06.2025 09:14 —
👍 21
🔁 3
💬 1
📌 0

Very much looking forward to the ChemBioChem and ChemPhysChem symposium "Advances in Structural Analysis of Biomolecules" in Berlin on October 23rd and 24th. If you are interested in the field, check out this conference: www.cbc-cpc2025.org (1/2)
26.05.2025 15:13 —
👍 7
🔁 2
💬 1
📌 0
Keynotes:
@machine.learning.bio Christian Dallago, Duke University, US
@delafuentelab.bsky.social Cesar de la Fuente, UPenn, US
@fragrisoni.bsky.social Francesca Grisoni - TU/e, NL
Birte Hoecker, Bayeruth University, DE
07.04.2025 10:27 —
👍 5
🔁 1
💬 0
📌 0
🧫Now in Bioinformatics Advances: "peptidy: A light-weight Python library for peptide representation in machine learning"
Find it here: https://doi.org/10.1093/bioadv/vbaf058
Authors include: @fragrisoni.bsky.social
04.04.2025 10:02 —
👍 2
🔁 1
💬 1
📌 0

We just preprinted a fresh study on molecular machine learning on OOD molecules 🧠
Using joint modeling, we could detect distribution shifts, estimate prediction reliability, and capture meaningful molecular patterns!
doi.org/10.26434/che...
#AI #chemistry
17.03.2025 14:52 —
👍 14
🔁 1
💬 0
📌 0
Very much looking forward to the 25th anniversary symposium of ChemBioChem and ChemPhysChem #CBCCPC2025 in Berlin on October 23rd and 24th, 2025.
There is an ever growing list of amazing speakers confirmed for this event. (1/2)
#chemsky #ChemBio @chemistryeurope.bsky.social
www.cbc-cpc2025.org
03.03.2025 13:57 —
👍 9
🔁 6
💬 3
📌 0

Working on #explainable #AI #ML for #chemistry and #materials? Then join us at the special track #xAI 2025 in Istanbul this July and submit your paper by Feb 28! xaiworldconference.com/2025/explain... Co-organised with @fragrisoni.bsky.social Pascal Friederich, Geemi Wellawatte etc.
24.02.2025 20:47 —
👍 10
🔁 1
💬 0
📌 0
Welcome to the other side!
11.02.2025 21:35 —
👍 1
🔁 0
💬 0
📌 0
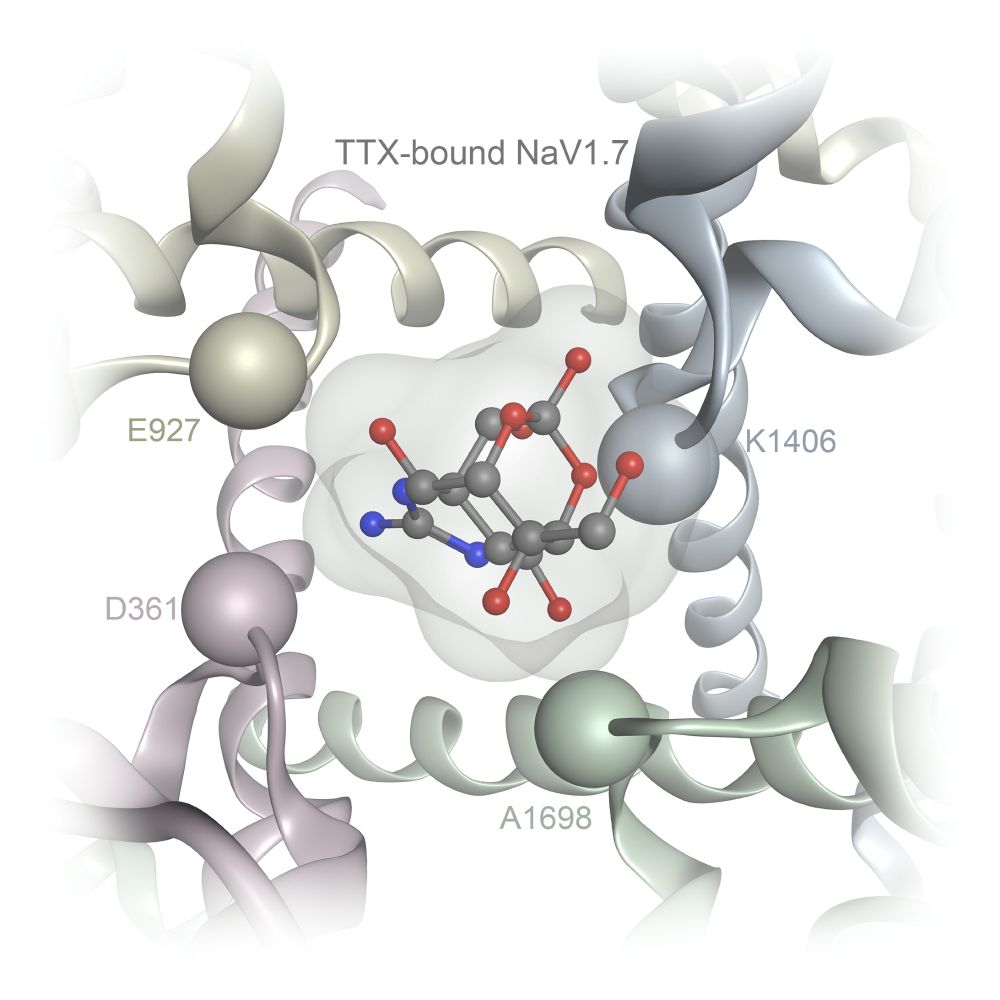
Very excited to share our review on "The Chemistry and Biology of the Tetrodotoxin Natural Product Family", where we discuss their potential for the development of analgesics, structure-activity relationships on NaV channels, biosynthetic hypotheses and chemical syntheses: tinyurl.com/ye6nwnwu.
30.01.2025 08:22 —
👍 17
🔁 2
💬 1
📌 0

The Jungle of Generative Drug Discovery: Traps, Treasures, and Ways Out
"How to evaluate de novo designs proposed by a generative model?" Despite the transformative potential of generative deep learning in drug discovery, this seemingly simple question has no clear answer...
If you use generative #DeepLearning for molecule design, check out our latest work, where we perform a large scale analysis (~1 B designs!) and find ‘traps’, ‘treasures’ and ‘ways out’ in the jungle of generative drug discovery.
🌴 🐒
Paper: arxiv.org/abs/2501.05457
Code: github.com/molML/jungle...
14.01.2025 20:29 —
👍 74
🔁 14
💬 0
📌 1
Hope never dies 😂
26.12.2024 17:49 —
👍 3
🔁 0
💬 0
📌 0
Please note that applications submitted via email and not through the dedicated portal will not be considered.
26.12.2024 15:09 —
👍 5
🔁 0
💬 1
📌 0
The surprising ineffectiveness of molecular dynamics coordinates for predicting bioactivity with machine learning
Authors: Emanuele Criscuolo, Rıza Özçelik, Derek van Tilborg, Francesca Grisoni
DOI: 10.26434/chemrxiv-2024-rp81v
18.12.2024 12:17 —
👍 4
🔁 1
💬 0
📌 0
Automated navigation of condensate phase behavior with active machine learning
Authors: Yannick Leurs, Willem van den Hout, Andrea Gardin, Joost van Dongen, Jan van Hest, Francesca Grisoni, Luc Brunsveld
DOI: 10.26434/chemrxiv-2024-frnj3
04.12.2024 05:11 —
👍 4
🔁 2
💬 0
📌 0
Open postdoc position available in my group in AI-assisted molecular simulations: https://www.rug.nl/about-ug/work-with-us/job-opportunities/?details=00347-02S000B1LP&cat=wp #aichem
28.11.2024 18:25 —
👍 19
🔁 12
💬 0
📌 0
Welcome Barbara! 👋
27.11.2024 20:39 —
👍 1
🔁 0
💬 1
📌 0
Thank you for attending! 🙂
26.11.2024 19:15 —
👍 1
🔁 0
💬 0
📌 0
Thanks @fragrisoni.bsky.social for a great talk at the University of Amsterdam! Learned a lot about how to apply AI effectively to molecular structure generation. Strong start to the #ChemAI events this week
26.11.2024 17:16 —
👍 3
🔁 1
💬 1
📌 0

Learning on compressed molecular representations
Last year, a preprint gained notoriety, proposing that a k-nearest neighbour classifier is able to outperform large-language models using compressed text as input and normalised compression distance (...
Fresh off the presses:
In "Learning on compressed molecular representations" Jan Weinreich and I looked into whether GZIP performed better than Neural Networks in chemical machine learning tasks. Yes, you've read that right.
TL;DR: Yes, GZIP can perform better than baseline GNNs and MLPs. It can ..
21.11.2024 12:58 —
👍 133
🔁 28
💬 4
📌 11
Hey Ben, ML for drug discovery here 🙂
15.11.2024 18:58 —
👍 7
🔁 0
💬 1
📌 0
🙋🏼
🙃
15.11.2024 18:50 —
👍 2
🔁 0
💬 0
📌 0