Themes:-
Session 1:- Trust the process
Session 2:- How to focus on the important details
Will you join the next one ?
Anish Kirtane
@dnasaur.bsky.social
eDNA Scientist at ETH Zurich
@dnasaur.bsky.social
eDNA Scientist at ETH Zurich
Themes:-
Session 1:- Trust the process
Session 2:- How to focus on the important details
Will you join the next one ?




Most days I do my science at the University, and my art at home. Sometimes, I get to bring the art with me to the University.
Great 2-day workshop with students from @ethz.ch and
@uzh-ch.bsky.social. I was amazed to see how naturally everyone was able to pick up watercolor techniques and vision.

Job offer: PostDoc in biotic and trophic interactions 90% (f/m/d) As part of the interdisciplinary PAPPUS project, which explores how plant communities and urban green space management support biodiversity and residents' quality of life, you will work with existing data. Your focus will be on analyzing biotic and trophic interactions between plants and arthropods, including predators, herbivores, pollinators, and detritivores. The data were collected from 60 public and private green spaces in Zurich. Your first task will be to analyse metabarcoding data from over 1200 invertebrate samples to generate a species lists. You will then examine the structure of mutualistic and antagonistic multitrophic networks, including food webs, and model their responses to urbanisation, management intensity, and plant preferences among residents. The position offers flexibility to develop your own research ideas within the scope of PAPPUS. You are expected to publish your findings in high-impact journals and present them at international conferences. Engagement with practitioners, especially through presentations and outreach in French, is also an important part of this PostDoc. You have a PhD degree in environmental science or in a related field. You have a good publication record as well as a background in ecological sciences and quantitative analyses. Proficiency in bioinformatic, metabarcoding, multitrophic ecology (food web), and ecological modelling is required, as is good knowledge of trait-based ecology. Practical and interdisciplinary experiences (e.g., with urban engineers) and knowledge of the principles and theories of urban ecology are helpful. You are ambitious, motivated and communicative. You are used to a high standard of independent, structured and you demonstrate a high level of team spirit. Proficiency in English and French, as well as good knowledge in German is required.
🌿 #PostDoc Alert! PhD in #EnvironmentalSciences? Proficient in #bioinformatic, #EcologicalModelling and good knowledge of trait-based #ecology🦋? Our #ConservationBiology group offers a PostDoc position in biotic and trophic interactions🐜: apply.refline.ch/273855/1769/... #UrbanEcology #ScienceJobs
20.10.2025 06:29 — 👍 12 🔁 9 💬 0 📌 0Unfortunately not, but would have been interesting to see that. Also just filtering 1L with 64 um would have allowed to separate the effects of volume and filter pore size !
11.03.2025 14:34 — 👍 2 🔁 0 💬 1 📌 0Thanks @rleonsampedro.bsky.social for the great picture !
02.03.2025 20:37 — 👍 2 🔁 0 💬 0 📌 0


We kicked off this semester's #SMEE with two great talks by @dnasaur.bsky.social and @phagemuffin.bsky.social. @usyseth.bsky.social
28.02.2025 17:28 — 👍 7 🔁 2 💬 1 📌 0Yeah, stuff like this keeps me up at night ...
21.02.2025 10:00 — 👍 1 🔁 0 💬 0 📌 0
🚀 Excited to share our new paper:
“Navigating Past Oceans: Comparing #Metabarcoding and #Metagenomics of Marine Ancient Sediment eDNA”
DOI: doi.org/10.1111/1755...
We dive into 8,000 years of marine life from Skagerrak sediments to show what different tools tell us about past #oceans! 🌊🧬

Big thanks to all the collaborators and co-authors @alpineedna.bsky.social ... See more detailed methods and a lot more analyses in the publication openly accessed via this link dx.doi.org/10.1002/edn3... (8/8)
21.02.2025 09:56 — 👍 0 🔁 0 💬 1 📌 0
Interestingly, these patters were different for ITS plant marker and COI metazoan marker! How does the source organisms impact spatial and particle size distribution of their eDNA? Can we optimise our capture methods depending on the target group? More work to be done! (7/8)
21.02.2025 09:56 — 👍 0 🔁 0 💬 1 📌 0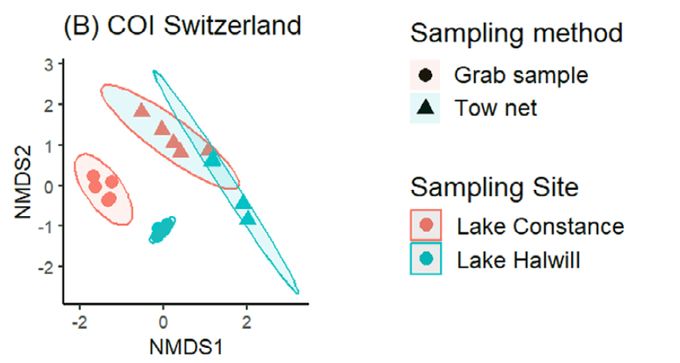
Not just the number but also the type of taxa differ. Beta diversity plots show samples cluster by location (as expected) but also by capture method ! Your choice of methods can dramatically impact your eDNA results ! (6/8)
21.02.2025 09:56 — 👍 0 🔁 0 💬 1 📌 0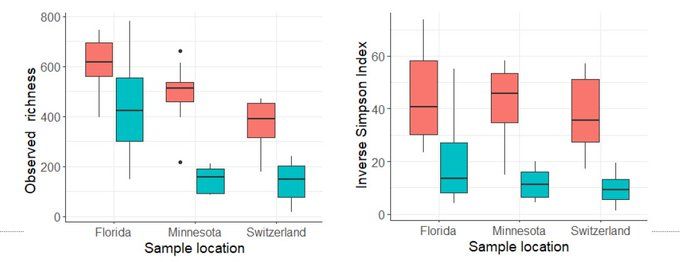
However, the metabarcoding results show more unintuitive patterns. Although the grab samples capture 100 times less DNA, the DNA it does capture represents significantly greater alpha diversity than that captured in tow nets. These results were consistent across all sites. (5/8)
21.02.2025 09:56 — 👍 1 🔁 0 💬 1 📌 0
We conducted the experiment in three different locations with varying water matrices: Switzerland, Montana, and Florida and found surprising consistent results ! First, we see that the tow nets (blue) captured ~100 times for DNA than grab sample filters (red). (4/8)
21.02.2025 09:56 — 👍 1 🔁 0 💬 1 📌 0
So here we tested whether increasing volume of water processed, indeed improved species diversity detection metabarcoding. We compared two extremes of volume-pore size combinations 1) 1L via 0.45 um filter and 2) 3500-7000 L filtered via 64 um tow net. (3/8)
21.02.2025 09:56 — 👍 0 🔁 1 💬 1 📌 0
The paper starts with a review of 300+ studies showing water volume depends on filter pore size. Thus, it may be intuitive to use larger pores to process more water and capture patchily distributed eDNA diversity. (2/8)
21.02.2025 09:56 — 👍 1 🔁 0 💬 1 📌 0
How, what, and where of environmental DNA #eDNA 🤔 Happy to share our newest publication exploring how volume of water processed and the pore size of the filter impacts the diversity of plant and animal communities captured via metabarcoding. (1/8)
onlinelibrary.wiley.com/doi/10.1002/...
The paper covers key topics in both fields, highlighting what biologists and hydrologists need to know about each other's work. We provide actionable steps on how they can integrate a multidisciplinary approach in their research. Shout out to all the co-authors and participants of the conference !
18.02.2025 10:15 — 👍 0 🔁 0 💬 0 📌 0
In this paper, we discuss how biologists using #eDNA in freshwater systems can benefit from integrating hydrological approaches to track the source of their eDNA, and how our understanding of water movement can be improved by using eDNA as a hydrological tracer (2/n)
18.02.2025 10:15 — 👍 1 🔁 0 💬 1 📌 0
Last year, biologists and hydrologists met at #eDNA4Hydro workshop in Bern, Switzerland to learn from each other and explore the intersection of our fields via eDNA. What we found was a new flow path (1/n) wires.onlinelibrary.wiley.com/doi/10.1002/...
18.02.2025 10:15 — 👍 9 🔁 6 💬 1 📌 0