
Excited to be in sunny San Diego for #NeurIPS2025 🌴
Biohub @czbiohub.bsky.social has a booth! Stop by to see cool demos from our computational imaging group and chat about AI for bioimaging.
@alxndrkalinin.bsky.social
AI/ML for cell image analysis @biohub.org | prev Broad Institute, CUHK-SZ & UMich

Excited to be in sunny San Diego for #NeurIPS2025 🌴
Biohub @czbiohub.bsky.social has a booth! Stop by to see cool demos from our computational imaging group and chat about AI for bioimaging.

A game changer in our hands... www.nature.com/articles/s41...
11.11.2025 18:48 — 👍 30 🔁 17 💬 1 📌 0
Janelia is hosting a course on DL for bioimage analysis www.janelia.org/you-janelia/...
06.11.2025 17:02 — 👍 36 🔁 13 💬 0 📌 0Today is the last chance to register for #CytoData2025. Don’t miss a fantastic program covering the full spectrum of image-based profiling!
cytodata25.eu-openscreen.eu
Shout out to the whole team: @drannecarpenter.bsky.social, @shantanu-singh.cc , @maom.bsky.social! 🙌
📖 Paper: openaccess.thecvf.com/content/ICCV...
💻 Code: github.com/alxndrkalini...
6/6

Plot showing acceleration of 3D cell segmentation CellProfiler tutorial pipeline from over 2 mins on CPU to less than 10 sec on GPU using cubic
We show how cubic can accelerate existing workflows by 10–1500×, including a 3D cell segmentation tutorial from CellProfiler. ⚡
5/6

Code snipper showing that unlike scikit-image/cuCIM, cubic can run the same code on either CPU or GPU based on the location of the input
cubic keeps things familiar: swap imports, put your image on the device you want, and the same function names automatically dispatch to the right CPU/GPU implementation—optional acceleration with minimal refactoring. ✨
4/6
CuPy-based cuCIM mirrors much of scikit-image, but uses device-specific function signatures that must match the input array’s device—typically leading to substantial refactoring to add GPU support to existing codebases.
3/6
scikit-image is widely used across bioimage analysis (incl. under the hood in CellProfiler), but with large 3D volumes and long time-lapse datasets, CPU execution often becomes the bottleneck.
2/6
💡Just presented our new paper at the @iccv.bsky.social BioImage Computing workshop: cubic: CUDA-accelerated 3D Bioimage Computing. We introduce a simple way to add GPU acceleration to scikit-image–based bioimage processing pipelines by swapping import statements. 🧵
1/6 #iccv2025

Large images have to be broken into tiles both for training and inference with neural networks. The tile predictions then need to be merged to produce the final volume prediction.
Segment large images without tiling artifacts: sharing our work that should have been presented at ICCV in 2 weeks - the brilliant first author Elena can’t go because of visa issues.
The paper: arxiv.org/abs/2503.19545 1/🧵
Morph Map is now published in Nature Methods. Excited to see what the community discovers with this resource mapping ~15,000 human genes!
rdcu.be/ezGre

Today @nature.com
www.nature.com/articles/d41...
In applications, there are going to be some fun CV presentations in the GenBio workshop - come check it out!
Disclaimer: I have one of those and would love deeper critique from a CV standpoint:
bsky.app/profile/alxn...
Workshop webpage: genbio-workshop.github.io/2025/
#ICML2025 #ICML #GenAI #GenBio #BioML #BioimageAnalysis

📄 Dive into the details in our preprint → arxiv.org/abs/2507.05383
I’ll be presenting this work at the GenBio workshop at ICML on Friday, July 18 — come say hi and chat about virtual staining!
Big cheers to our collaborators at @imbavienna.bsky.social & @umich.edu Medical School
5/5

The visual difference is clear: compared to a baseline (F-net), Spotlight sharply reduces artifacts, resulting in clearer nuclear boundaries and less segmentation artifacts, while preserving foreground textures.
4/5
💡Spotlight uses the fact that even simple histogram thresholding (e.g., Otsu) is often sufficient to approximate informative FG regions. We use this to (1) mask MSE loss to focus learning on FG intensities, and (2) add a FG/BG segmentation loss to preserve cell morphology.
3/5
Most VS models are trained with pixel-wise losses like MSE, treating background (BG) and foreground (FG) equally. Unlike natural images, BG in cell imaging isn't informative–so models learn to reproduce noise. E.g., in 3D, predictions show axial blur and elongation.
2/5

Figure 1:A schematic overview of Spotlight. a: Typical virtual staining models are trained using a pixel-wise loss such as mean squared error (MSE) on the whole image. b: Spotlight uses foreground estimation obtained by histogram thresholding to restrict pixel-wise loss to foreground areas and also employs soft-thresholding of the prediction to compute segmentation loss.
🔬🤖 Introducing Spotlight: virtual staining (VS) improved by focusing on cells
VS models often learn to predict both cells and noisy background, because training treats all pixels equally. We address this by explicitly training models to prioritize foreground.
1/5
Looks neat! How does the number/variety of features compare to Cellprofiler? Does it have Python bindings?
09.07.2025 17:18 — 👍 0 🔁 0 💬 1 📌 0Big shout out to the whole team: Alán F. Muñoz, Tim Treis, @shatavishadg.bsky.social , @fabiantheis.bsky.social ntheis.bsky.social, @drannecarpenter.bsky.social, @shantanu-singh.cc
6/6

We will be discussing this work at #ICML2025 CODEML workshop - come check it out and chat with us!
📖 Arxiv → arxiv.org/abs/2507.01163
💻 Github → github.com/afermg/cp_me...
5/6
Key benefits:
- Reproducibility: replaces GUI workflows with code
- General: agnostic to data types (3D images, spatial transcriptomics)
- Few dependencies: easy to integrate into existing image analysis frameworks
- Backwards-support: largely matches CellProfiler features
4/6
With that in mind, we developed cp_measure, a Python library that extracts morphological features from segmented images from within your pipeline, bridging the gap between the BioAI/ML community and the existing GUI-based tool that populates bioimaging workflows.
3/6
We felt there were a limited number of programmatic tools for featurizing segmented cell images, and CellProfiler is the de-facto standard for interpretable features.
2/6
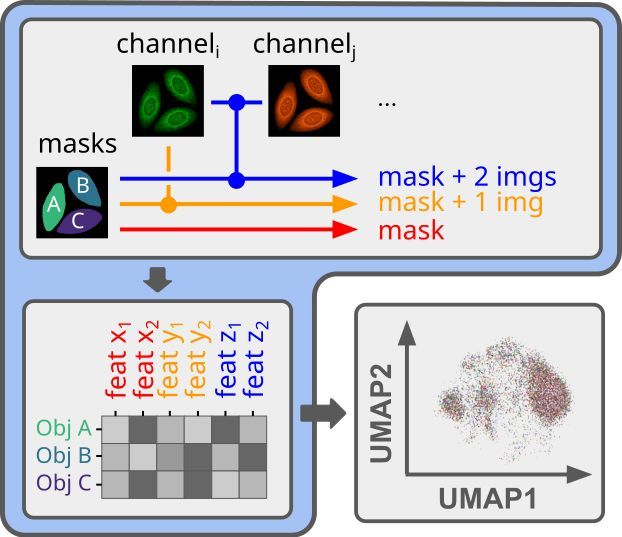
🔬API-first feature extraction for image-based profiling workflows
If you need to obtain interpretable features from your segmented microscopy images, but want to do it in a fully automated way, we know the struggle.
1/6
If you’re interested in single cell data analysis, come give Image based profiles a try! Huge dataset being made available for exploration at this hackathon (+ symposium):
Berlin, November cytodata25.eu-openscreen.eu

The 2nd AI4Life Challenge is live!
Calling the AI & bioimaging community to tackle a key microscopy challenge: removing noise while preserving detail.
📦 Paired noisy/clean datasets
📈 Ground-truth evaluation
🧠 DL focus
Build, test, compete 👉 ai4life.eurobioimaging.eu/challenge-2/
There is still time to submit an abstract to CytoData 2025!
23.06.2025 16:36 — 👍 1 🔁 0 💬 0 📌 0