Thanks Maria!
30.10.2025 15:51 — 👍 0 🔁 0 💬 0 📌 0Assaf Zaritsky
@assafzaritsky.bsky.social
Computational cell biologist https://www.assafzaritsky.com/
@assafzaritsky.bsky.social
Computational cell biologist https://www.assafzaritsky.com/
Thanks Maria!
30.10.2025 15:51 — 👍 0 🔁 0 💬 0 📌 0Somehow missed acknowledging Alon Shpigler, the first author 😱
30.10.2025 14:15 — 👍 0 🔁 0 💬 0 📌 0
Congratulations to Naor Kolet who led this project and thanks to co-authors Shahar Golan for his contribution and to @erinweisbart.bsky.social l for her critical insights regarding interpretability 🎉
3/3
Anomaly-based representations have several benefits:
1. Reducing batch effects
2. Improving reproducibility and mechanism of action classification
3. Complementing classical representations
4. Revealing alterations in biologically interpretable dependencies
2/3

Our paper is out ✨
doi.org/10.1016/j.ce...
We propose a self-supervised anomaly representation that encodes morphological inter-feature dependencies for high-content image-based cell phenotyping!
1/3
Join us!
10.10.2025 14:37 — 👍 0 🔁 0 💬 0 📌 0Jennifer Lippincott-Schwartz, Emma Lundberg, Alex Mogilner, Maddy Parsons, @loicaroyer.bsky.social, Guillaume Salbreux, Anđela Šarić, Timm Schroeder, Hervé Turlier, @virginieuhlmann.bsky.social, Vincenzo Vitelli
Please save the date and spread the word!
3/3
Co-organized with Meghan Driscoll, @quantumjot.bsky.social and Susanne Rafelski
Speakers include: @kevin-dean.bsky.social, @ellenberglab.bsky.social, Margaret Gardel, @florianjug.bsky.social, @krishnaswamylab.bsky.social, @levayerr.bsky.social, @priscaliberali.bsky.social, ...

Excited to announce the first @cshlnews.bsky.social meeting on Cell Modeling in Space and Time, June 2026 💫
meetings.cshl.edu/meetings.asp...
Bringing together experiments, computation and theory to explore dynamic, multi-scale cellular organization and function!
1/3

Nature Biomedical Engineering is hiring again! This time we're hoping to add an editor with machine learning expertise to the team (although we are open to those with other relevant expertise!) Deadline is Oct 20. Please RT! springernature.wd3.myworkdayjobs.com/SpringerNatu...
24.09.2025 18:36 — 👍 17 🔁 18 💬 1 📌 2
📢We're #hiring Group Leaders!
Apply to lead a lab at Janelia & advance biology using theory, computational modeling & machine learning.
🔹5-year renewable appointment
🔹Pioneer new tools & approaches
🔹Collaborate across disciplines
Apply by Nov. 4👉 https://janelia.link/groupleader
We are searching for an enthusiastic scientist to join our team and establish annual killifish as a model system for early developmental biology and biophysics! @embl.org @embldbunit.bsky.social
Apply here:
embl.wd103.myworkdayjobs.com/EMBL/job/Hei...
Please re-post 🙏
How do organelles change over the course of a stem cell’s life?
ADIs @cohenlaboratory.bsky.social, @assafzaritsky.bsky.social , and @mattersoflight.bsky.social are using ML and cell engineering to explore the #FrontierScience of stem cell differentiation.
alleninstitute.org/division/fro...

BIG start to #mmc2025 with Assaf Zaritsky giving an awesome opening plenary.
@royalmicrosoc.bsky.social @eurobioimaging.bsky.social @globias.bsky.social @bioimaginguk.bsky.social @globalbioimaging.bsky.social
Sorry, I meant @leeat-keren.bsky.social and Yael Amitay....
09.05.2025 09:15 — 👍 0 🔁 0 💬 0 📌 0
A creative and technically challenging idea led by @zamir_amos, with key contributions from Yuval Tamir and @yaelam75. This would not have been possible without the great collaboration with @leeat_keren! And thanks to @WellcomeLeap ΔTissue for funding!
16/n
CISM source code is publicly available github.com/zaritskylab/...
Also, check CISM’s backbone FANMOD+, that enables efficient motifs extraction in “multi-colored” networks github.com/zaritskylab/...
Try it out!
15/n

Our results suggest that the local organization of a few cells in discriminative motifs are emergent properties that may define an intermediate spatial scale driving tissue function
14/n

CISM’s unsupervised motif enumeration followed by supervised context-dependent selection distils the discriminative motifs from the huge and noisy landscape of all putative sub-networks of intercellular interactions and provides discrimination along with interpretability
13/n
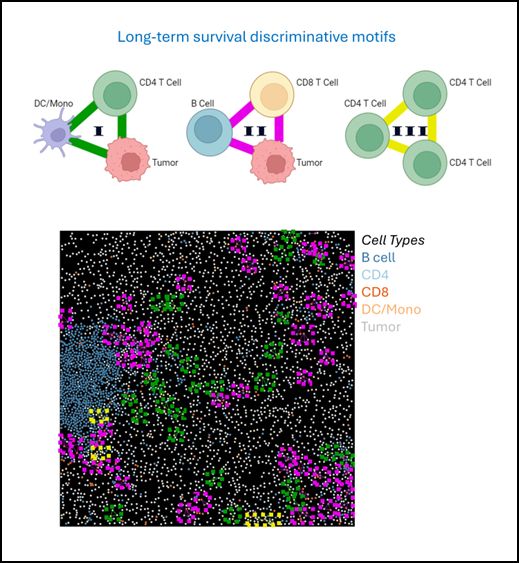
To demonstrate the general applicability of CISM for identifying local cellular structures linked to disease states, we applied it to investigate the tumor microenvironment in a human cohort of Breast Cancer (TNBC) patients, comparing short-term and long-term survivors.
12/n

The same motifs can be differentially analyzed according to different context-dependent selection of the discriminative motifs. We demonstrate this by investigating the immune microenvironment of NP versus PN (metastatic lymph nodes that did not develop distant metastases)
11/n

Spatial interpretation of motifs localization patterns reveals an association between (local) motifs to (global) tissue compartments highlighting the potential contribution of CISM to multi-scale analysis and interpretation
10/n

The spatial arrangement of cell types within the discriminative motifs contributed to disease state classification, indicating that the specific intra-motif cell-cell interactions are sensitive markers for disease state beyond their cell type composition
9/n

Exploring the landscape of the discriminative motifs-induced cell distribution and motifs-induced pairwise cell-cell interactions revealed differential composition of cell type and cell-cell interactions
8/n

Classifying NN vs. NP disease states using discriminative four-cell motifs outperformed other methods, suggesting that these motifs can act as multicellular signatures of disease
7/n

We applied CISM to investigate future metastases by analyzing the immune microenvironment in tumor-free lymph nodes of melanoma patients using multiplexed imaging.
Cohort and data by our collaborators @yaelam75 and @leeat_keren are described here doi.org/10.1101/2024...
6/n

These discriminative motifs’ representations can be interpreted based on their prevalence, contribution to classification, identity of the cells that comprise them and their pairwise interactions and their spatial location in respect to more global tissue compartments
5/n

CISM context-dependent discriminative motifs can be used to formulate a machine learning-based prediction of the tissue’s disease state where each patient is represented by the (sparse) vector of its discriminative motifs' frequencies
4/n

CISM reduces the number of potential high-order interactions with unsupervised selection of ‘motifs’, enriched local multicellular structures, and then associates these motifs with the tissue disease state according to their presence in patients at different clinical states
3/n

Source: https://doi.org/10.15252/msb.202110726
The clinical manifestation of diseased tissue arises from intricate intercellular interactions that extend beyond pairwise cell-cell interactions. Deciphering these processes is challenging due to the combinatorial complexity of multicellular organization.
2/n