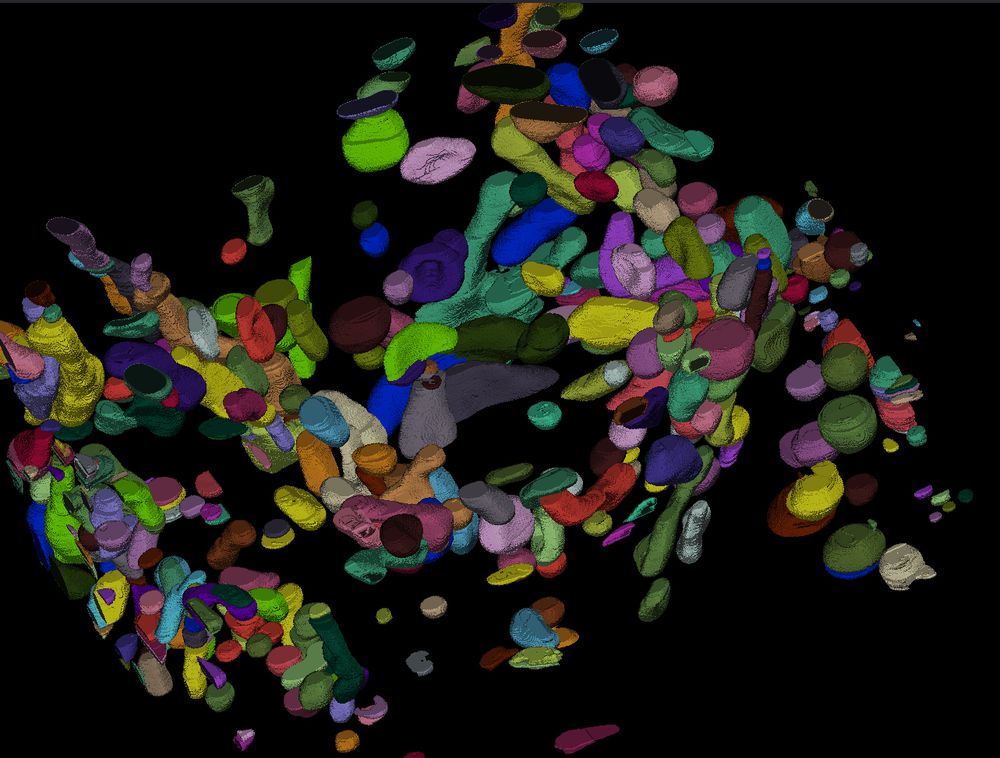
Rendering of a full cochlea (left) with three stains (PV, VGlut3, CTBP2) shown in three different colors (red, blue, cyan). The whole cochlea is a spiral shaped structure, with spiral ganglion neurons (SGNs) in the inner helix, labeled by PV and inner hair cells (IHCs) in the outer helix, labeled by Vglur3. The figure also shows zoom ins. The right hand side shows segmentation results for SGNs, IHCs (represented by colored masks) and synapse detections (represented by colored dots).
Preprint alert! CochleaNet, our framework for analyzing light-sheet data of the cochlea. It consists of three networks to segment spiral ganglion neurons, inner hair cells, and to detect synapses. See rendering of a full cochlea in the image, find the preprint at doi.org/10.1101/2025....
18.11.2025 07:58 — 👍 26 🔁 4 💬 1 📌 1

Don’t miss Elena’s (from @ilastik-team.bsky.social lab) brilliant work @ #ICCV2025 with @anwaiarchit.bsky.social & @cppape.bsky.social poster 292 @ 11:15AM 🔬They tackle segmentation of massive 3D microscopy images & show how BatchRenorm removes tiling artifacts boosting transferability and clarity🌺
22.10.2025 20:23 — 👍 8 🔁 3 💬 0 📌 0

SynapseNet is a deep learning based software tool that automates the segmentation of vesicles, mitochondria, synaptic compartments, and the active zone. This is visualized in three panels. On the left, a section of an electron tomogram with a synaptic compartment densely filled with vesicles, which appear as round structures with dark boundary and light body in the image, is shwon. The top right panel shows the segmentation result of vesicles, visualized by masks with an individual color per vesicle and outlines for the segmented compartment (red) and active zone (blue). The bottom right panels shows a 3D rendering of the segmentation with vesicles shown as yellow spheres.
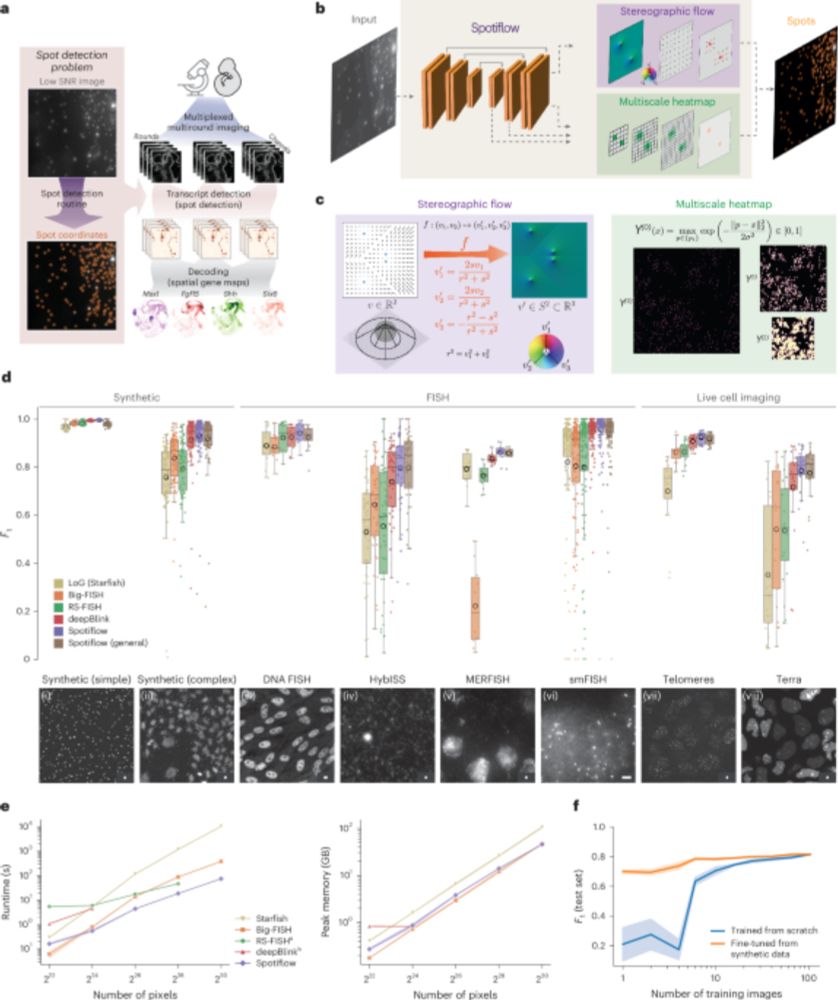
Are you studying synapses in electron microscopy? Tired of annotating vesicles? We have the tool for you! SynapseNet implements automatic segmentation and analysis of vesicles and other synaptic structures and has now been published:
www.molbiolcell.org/doi/full/10....
17.10.2025 11:09 — 👍 39 🔁 13 💬 2 📌 0

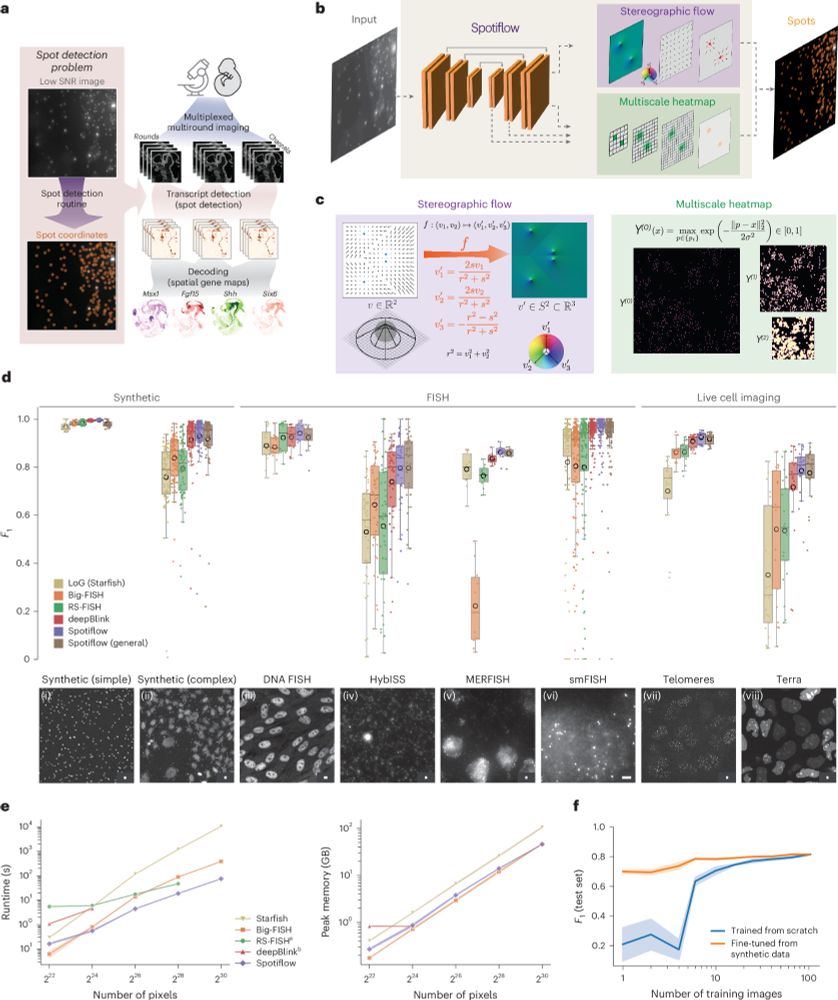
Large images have to be broken into tiles both for training and inference with neural networks. The tile predictions then need to be merged to produce the final volume prediction.
Segment large images without tiling artifacts: sharing our work that should have been presented at ICCV in 2 weeks - the brilliant first author Elena can’t go because of visa issues.
The paper: arxiv.org/abs/2503.19545 1/🧵
09.10.2025 12:56 — 👍 57 🔁 16 💬 2 📌 0
Calling all data - Nature Methods
As life sciences research becomes enmeshed in the age of AI, real experimental data are more valuable than ever.
Data is the key to AI advances in biology and "Still, when it comes to data, nothing compares to the real thing." An important editorial in Nature methods with a nice little shout out to microSAM:
www.nature.com/articles/s41...
07.08.2025 16:05 — 👍 5 🔁 1 💬 1 📌 0
Anwai represented the lab at MIDL very well! Read his thread for details on our two latest papers on foundation models for microscopy, histopathology and medical imaging.
16.07.2025 19:54 — 👍 10 🔁 1 💬 0 📌 0
There are exciting opportunities and foreseeable clinical applications of vision foundation models for biomedical image analysis! The MIDL Community did seem excited, and so are we! (some amazing follow-up applications coming soon, keep an eye open 😉)
16.07.2025 17:46 — 👍 0 🔁 0 💬 0 📌 0
And both of our efforts and these beautiful presentations at #MIDL2025 have been successful due to immense efforts from our amazing lab members @caroteu.bsky.social and Titus Griebel. And of-course @cppape.bsky.social being the absolute best PI, as always (I can't say this enough tbh)!
16.07.2025 17:46 — 👍 1 🔁 0 💬 1 📌 0

And sticking to our @cppape.bsky.social lab's theme of rocking in the open-source world, all the models and code are completely open to use and easy to access. You can take @midl-conference.bsky.social's word on this! 😉
PathoSAM: github.com/computationa...
Late PEFT: github.com/computationa...
16.07.2025 17:46 — 👍 1 🔁 0 💬 1 📌 0
Parameter Efficient Fine-Tuning of Segment Anything Model for Biomedical Imaging
Segmentation is an important analysis task for biomedical images, enabling the study of individual organelles, cells or organs. Deep learning has massively improved segmentation methods, but challenge...
Next, we introduce a novel finetuning paradigm for foundation models "Late PEFT", inspired by our striking observations of PEFT's memory requirements, questioning: "Are PEFT methods Resource Efficient?"
We have an answer: PEFT "can be resource efficient"! Check out more at: doi.org/10.48550/arX...
16.07.2025 17:46 — 👍 0 🔁 0 💬 1 📌 0


To begin with, here's "PathoSAM", our foundation model for nucleus segmentation in histopathology, presented as an Oral! (and my very first one).
If you want a SOTA model for interactive and automatic segmentation for your histopathology images, go check out the paper now doi.org/10.48550/arX...
16.07.2025 17:46 — 👍 1 🔁 0 💬 1 📌 0

We presented our latest work on "PathoSAM" and "Late PEFT" last week at #MIDL2025 (Salt Lake City)!
The community is growing and MIDL is becoming the venue-to-go for high quality research discussion!🧵
16.07.2025 17:46 — 👍 6 🔁 0 💬 1 📌 1
Are you looking for an exciting position at the intersection of super-resolution microscopy and AI? Then check out the PhD and PostDoc position we offer for a joint project with the Group of Stephan Hell at MPI Göttingen. Please share with anyone interested, read on for links and details.
07.07.2025 11:46 — 👍 19 🔁 11 💬 1 📌 1
Congratulations @maweigert.bsky.social and @albertdm.bsky.social 🥳
08.06.2025 16:44 — 👍 2 🔁 0 💬 0 📌 0
Announcing the new release v1.4.0 of microSAM. The main changes are:
1. Simplified installation on windows.
2. Preliminary support for automatic tracking.
3. Improved interface for model selection.
Read on for a quick summary of the changes.
28.03.2025 19:08 — 👍 18 🔁 2 💬 2 📌 0


@anwaiarchit.bsky.social & @cppape.bsky.social demonstrating the power of micro-sam, the napari plugin for the microscopy segment anything model, in their awesome workshop at the #TiM2025 conference in Münsingen. 🔬🦠💻
19.03.2025 09:08 — 👍 18 🔁 3 💬 0 📌 0

Our March issue is now live! 🥳
nature.com/nmeth/volume...
The cover represents the process of cell and organelle segmentation by Segment Anything for Microscopy.
Paper here: nature.com/articles/s41...
Cover by Sebastian von Haaren.
13.03.2025 15:53 — 👍 18 🔁 8 💬 1 📌 1
Another feather for Segment Anything for Microscopy. We made it to the cover for @naturemethods.bsky.social!
And all thanks to our amazing @haarensv.bsky.social for this! <3
13.03.2025 16:25 — 👍 9 🔁 0 💬 0 📌 0
Hi @psobolewskiphd.bsky.social,
It's under the same name in our GUI model list. With the latest version installed, this will be stored in cache automatically!
03.03.2025 17:35 — 👍 2 🔁 0 💬 0 📌 0

Pflanzenzellen, die mit einem Fluoreszenzmikroskop aufgenommen und mit dem Modell automatisch segmentiert wurden. Die zugrunde liegenden Daten sind dreidimensional und das Bild zeigt eine Darstellung der segmentierten Zellen, die jeweils durch eine andere Farbe repräsentiert werden.
Foto: Nature Methods: 10.1038/s41592-024-02580-4

Segmentierung von Zellen in der Lichtmikroskopie mit μSAM. Das Bild zeigt, wie Zellen in der Phasenkontrastmikroskopie mit μSAM segmentiert werden können. Grüne Punkte und Kästchen zeigen die Benutzereingabe und farbige Masken die entsprechende Vorhersage des Modells.
Foto: Erstellt von Anwai Archit mit dem μSAM-Tool, verfügbar in Nature Methods: 10.1038/s41592-024-02580-4
Automatische Zellanalyse mit #KI: Forschende trainierten eine bestehende, KI-basierte Software neu. Das Modell „Segment Anything for Microscopy“ kann Bilder von Geweben, Zellen und anderen Strukturen genau segmentieren: s.gwdg.de/HqkMz2; s.gwdg.de/iTejKW
Forschungsteam mit bsky.app/profile/cppa...
26.02.2025 10:41 — 👍 11 🔁 3 💬 0 📌 0
It was soooo much fun to brainstorm solutions with everyone, together!❤️
#EMBLDeepLearning
24.02.2025 21:49 — 👍 3 🔁 0 💬 0 📌 0
Thank you @unigoettingen.bsky.social for the feature!😍
μsam got some super cool feature updates last week. Don't wait for the next release, go check us out now!
github.com/computationa...
24.02.2025 11:56 — 👍 4 🔁 0 💬 0 📌 0
micro_sam API documentation
Because we have seen these improvements and due to popular demand, cc @ritastrack.bsky.social @jianxuchen.bsky.social, we have decided to start a call for community data submission to further improve our models: computational-cell-analytics.github.io/micro-sam/mi... . Looking forward to any feedback
21.02.2025 10:36 — 👍 11 🔁 4 💬 1 📌 2


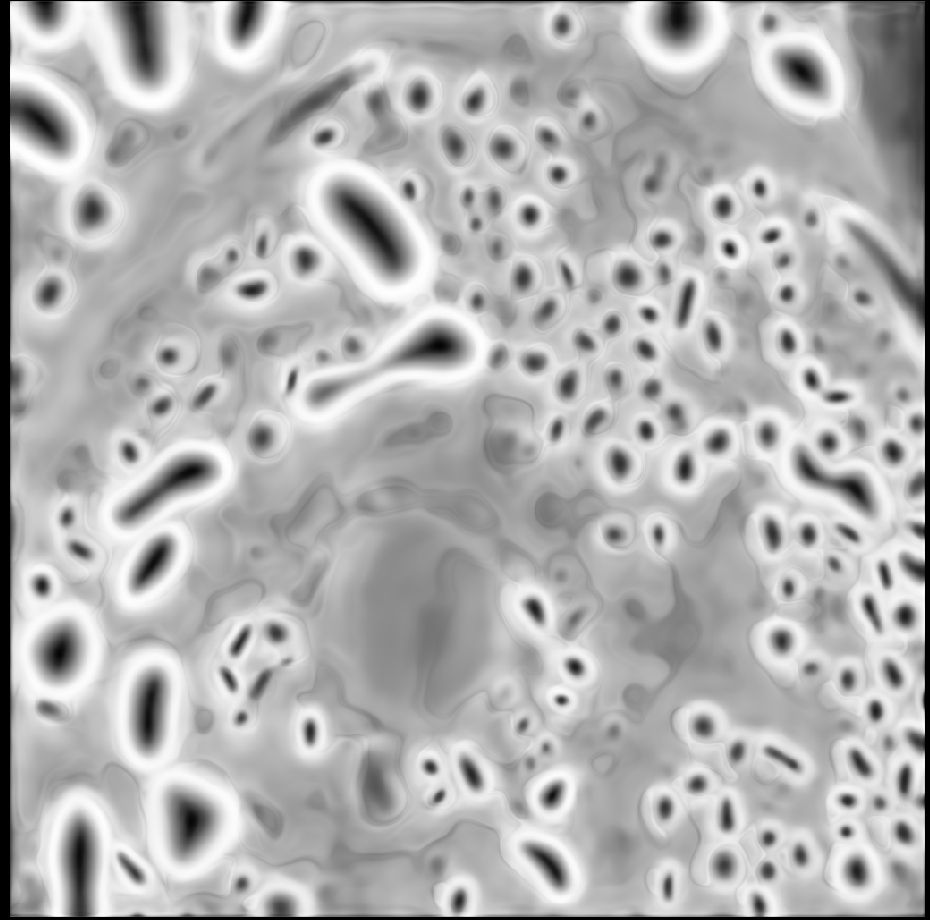
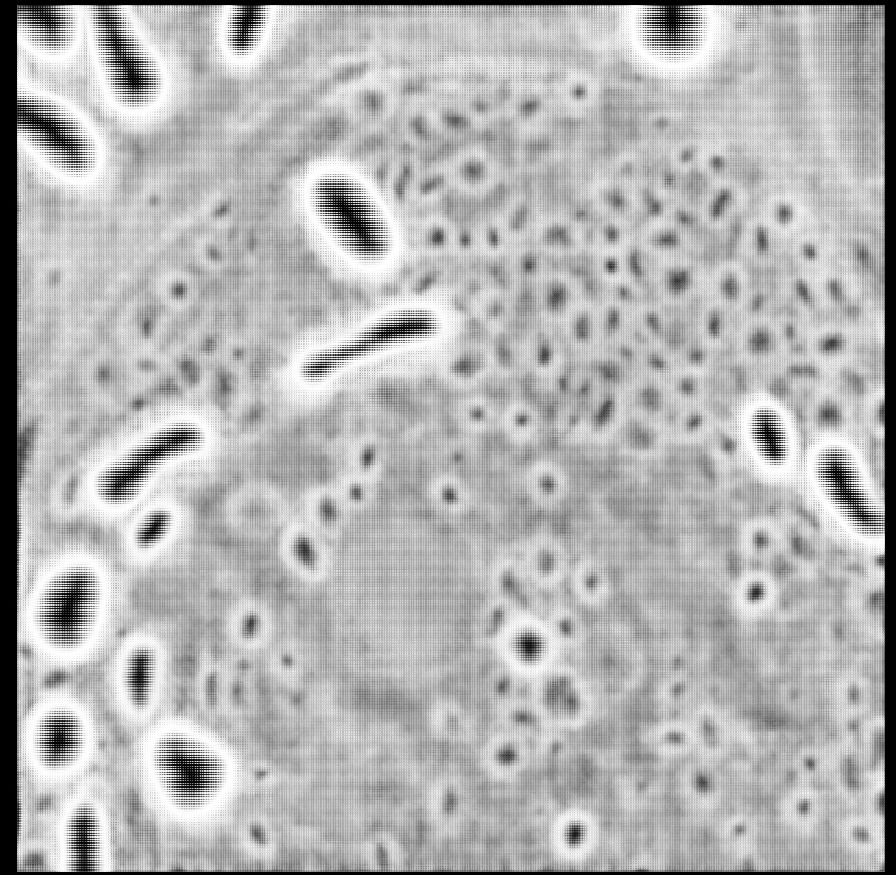
Our next micro_sam release is here! We have a new model for light microscopy, that massively improves for automatic segmentation! See the qualitative and quantitative comparison in the images, v2 is our previous version, v3 is the new one.
21.02.2025 10:36 — 👍 33 🔁 10 💬 3 📌 0
Help improve MicroSAM!
21.02.2025 14:08 — 👍 11 🔁 3 💬 0 📌 0
Look at all of those #OMEZarrs! 🤩
21.02.2025 10:22 — 👍 16 🔁 5 💬 0 📌 0
I actually tested these two and also SamCell (recently on Biorxiv). Here's the quick and dirty results - might still be finetuned, and I might have not always picked the best settings.
16.02.2025 21:29 — 👍 14 🔁 4 💬 1 📌 0
Research Software Engineer in bioimaging // I run the Human Organ Atlas (@humanorganatlas.bsky.social) // cyclist // walker
AI x Bioimaging @biohub.org | prev Broad Institute, CUHK-SZ & UMich
computation biology, drug discovery, computer vision, microscopy, statistics, machine learning, all happening at https://carpenter-singh-lab.broadinstitute.org/
Director, Max Planck Institute for Intelligent Systems; Chief Scientist Meshcapade; Speaker, Cyber Valley.
Building 3D humans.
https://ps.is.mpg.de/person/black
https://meshcapade.com/
https://scholar.google.com/citations?user=6NjbexEAAAAJ&hl=en&oi=ao
AIxBio Research Scientist 👩🔬 | PhD Computational Biology 👩💻 | BSc Biomedicine 🧫 | "Singlecellologist" 🦠 into biologically meaningful representation learning 🧬 | Decoding life in London 🇬🇧
Y. Eva Tan Professor in Neurotechnology, MIT. Investigator, HHMI. Leader, Synthetic Neurobiology Group, http://synthneuro.org. Scientist, inventor, entrepreneur.
Scientist, Group Leader @EMBL Heidelberg.
Research Scientist in Cell and Computational Biology at Helmholtz Munich | Creator of Cell-ACDC and SpotMAX | Excited about mitochondrial DNA and AI-driven bioimage analysis | Dad of 2
Scientist @ INSERM Institut de la Vision 🇫🇷 | Microscopy 🔬 | Transcriptomics 🧬 | Developmental Biology in 4D 🌟 - Ex Chan Zuckerberg Biohub SF 🇺🇸 / RIKEN 🇯🇵
Neuroscience, NeuroAI | https://kfranke.com/ | Senior Scientist at Stanford working at https://enigmaproject.ai/
Postdoc at the University of Göttingen (@huiskenlab), a physicist with an interest in Light-sheet microscopy, computer science, and developmental biology.
📝 https://www.nature.com/articles/s41587-025-02882-8
Cross-posting from idlethumbs.social/@ratamero
half respectable professional (life sciences data stuff), half nerdy gremlin. views my own etc etc. he/him/ele
https://erickratamero.com
Neuroscience & functional ultrasound imaging. Vision and brain states. Professor at University Medical Center Göttingen. https://brainwidenetworks.uni-goettingen.de/ Co-Spokesperson, EKFZ Center for Optogenetic Therapies. https://ekfz.uni-goettingen.de/en/
High-end modular and portable #lightsheet microscopy for inside and outside the lab / Flamingo project / Imaging development, physiology and anatomy in live specimens and cleared tissues. Former @morgridgeinstitute.bsky.social, @mpi-cbg.de
huiskenlab.com
A scientific journal publishing cutting-edge advances at the intersection of the life and physical sciences. Posts by the editors.
Chief Editor at Nature Biotechnology, running and science fanatic
Cell biologist | microscopist studying the nuclear and cell division of Malaria parasites | https://www.absalonlab.com/ |Co-founder of Peers in Parasitology | Be who you are 🏳️🌈 |Healthcare is a human right