
I'm hiring a computational biologist interested in complex trait genetics using deep learning approaches. Reach out to me, if interested.
12.09.2025 19:00 — 👍 38 🔁 46 💬 1 📌 0@skimhellmuth.bsky.social
Human Geneticist, Emmy Noether Group Leader at Helmholtz Munich & Dr. von Hauner Children‘s Hospital, University Hospital LMU Munich, Germany, Member of Die Junge Akademie

I'm hiring a computational biologist interested in complex trait genetics using deep learning approaches. Reach out to me, if interested.
12.09.2025 19:00 — 👍 38 🔁 46 💬 1 📌 0Excited to share we’ve been awarded the ERC StG 🎉 for our project ProSocial, studying neural mechanisms of prosocial behavior in health & disease. Huge thanks to my group for their amazing work & to great colleagues for their support! Positions opening soon—reach out if interested!
04.09.2025 14:20 — 👍 25 🔁 5 💬 7 📌 0Thrilled to share our lab's first preprint on the neural mechanisms underlying helping behavior in mice! 🧠🐭 We show that dorsal hippocampal networks play a key role in rescuing a distressed peer: www.biorxiv.org/content/10.1...
19.04.2025 16:06 — 👍 17 🔁 6 💬 0 📌 0
1. 🚨New preprint: tinyurl.com/tenk10k-causal.
We explored causal effects of gene expression in immune cell types on complex traits and diseases by combining single-cell expression quantitative trait loci (sc-eQTL) mapping in 5M+ cells from 1,925 donors in TenK10K study and GWAS. 🧵

New preprint alert: tinyurl.com/tenk10k-multiome. Excited to share our analysis on the impact of genetic variants on single-cell chromatin accessibility in blood, using scATAC-seq and WGS from over 1,000 donors and 3.5M nuclei as part of TenK10K phase 1 🧬
🧵👇 (1/n)
Excited to see that our study on context-specific transcription factors was named as one of 8 remarkable outputs of 2024 by the Swiss Institute of Bioinformatics.
05.08.2025 20:51 — 👍 4 🔁 2 💬 0 📌 0Great career opportunities in Cellular Genomics @sangerinstitute.bsky.social
Big data (human in vivo + in vitro) + AI to derive biological mechanisms at scale.
We are seeking a range of research expertise including spatial omics and perturbation at scale

🥳🌟🥳 Our very first PhD student defended her thesis yesterday!
Huge congratulations to 💫Dr. Sathya Darmalinggam💫 for a brilliant PhD defence 🎓!
It's been a privilege have Sathya in the lab and see her develop as an independent researcher, the lab and I couldn't be more proud🤩! #proudPI #WomenInSTEM
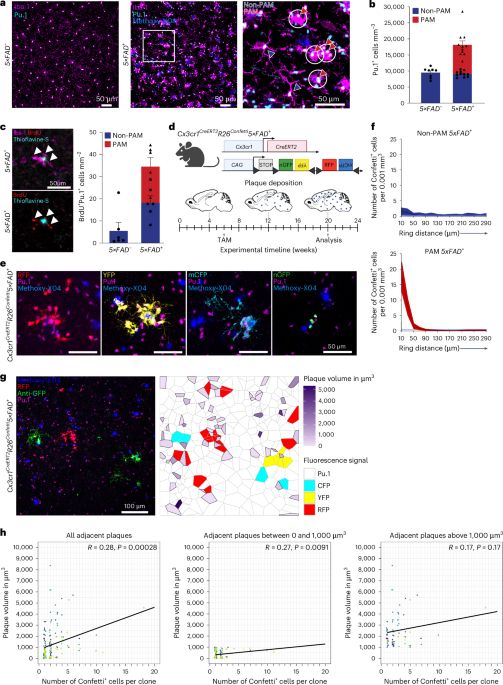
🥳🥳🥳Countless years in the making and there it is 🫶🏻
Exceptionally proud of my lab and especially Lance who was driving this project for the last years! 🏎️Thank you to all the fantastic collaborators ♥️ I hope everyone enjoys reading our little story! www.nature.com/articles/s41...
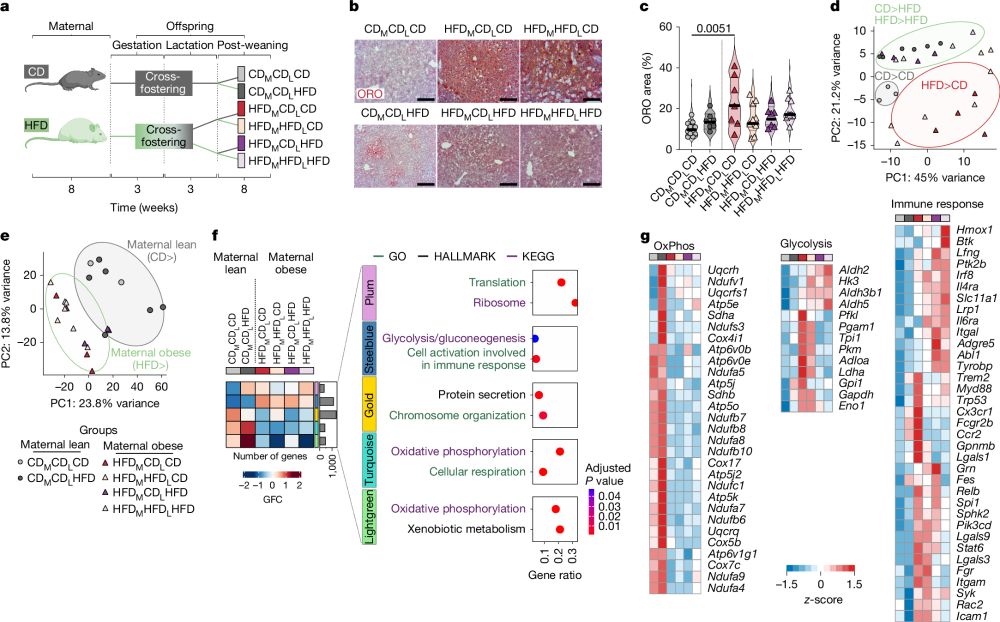
We are SO EXCITED to share our story @nature.com:
🧵1/
Maternal obesity reprograms fetal liver macrophages, triggering adult fatty liver disease in the offspring.
This study uncovers how Kupffer cells act as intergenerational messengers in metabolic disease.
👉 Read here: rdcu.be/erD3b

Preprint on "Improving spliced alignment by modeling splice sites with deep learning". It describes minisplice for modeling splice signals. Minimap2 and miniprot now optionally use the predicted scores to improve spliced alignment.
arxiv.org/abs/2506.12986

📢 Just posted: Our preprint introducing SPC — Spectral Components — is now live on medRxiv!
Led by Dr. Ruhollah Shemirani and years in the making, this method offers a robust, scalable way to adjust for recent population structure in genomic analyses.
🔗 www.medrxiv.org/content/10.1...
1/9

As a post-GWAS analysis, ever thought about automatic haplotype testing @ each locus + test vs any number of traits?
👏 Amazing @dariushghasemi.bsky.social just released #Haplomics, a #Snakemake pipeline walking u all the way down to summary stats+clustering
www.medrxiv.org/content/10.1...
#statgen

If we want to connect genetic variation to disease, we need to move beyond baseline conditions. Our study shows that dynamic, context-specific regulation holds the key to understanding many unexplained GWAS signals. The preprint again:
www.biorxiv.org/content/10.1...

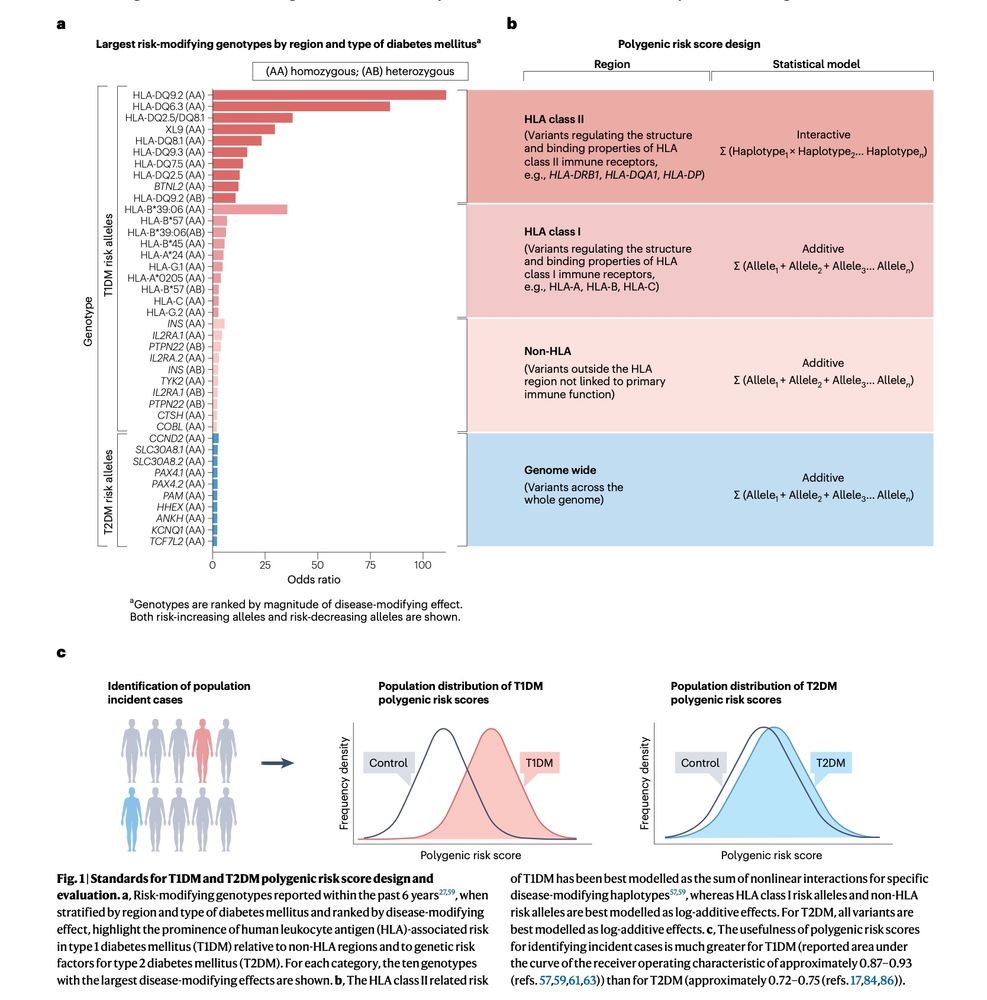
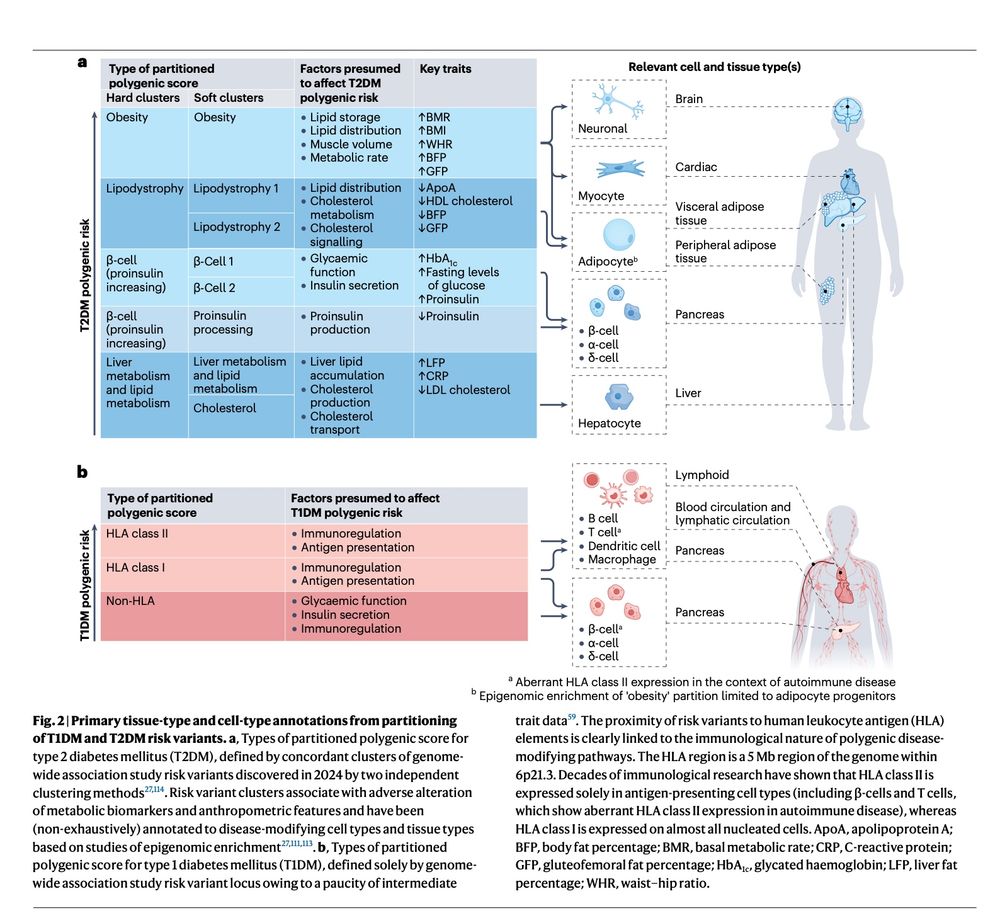
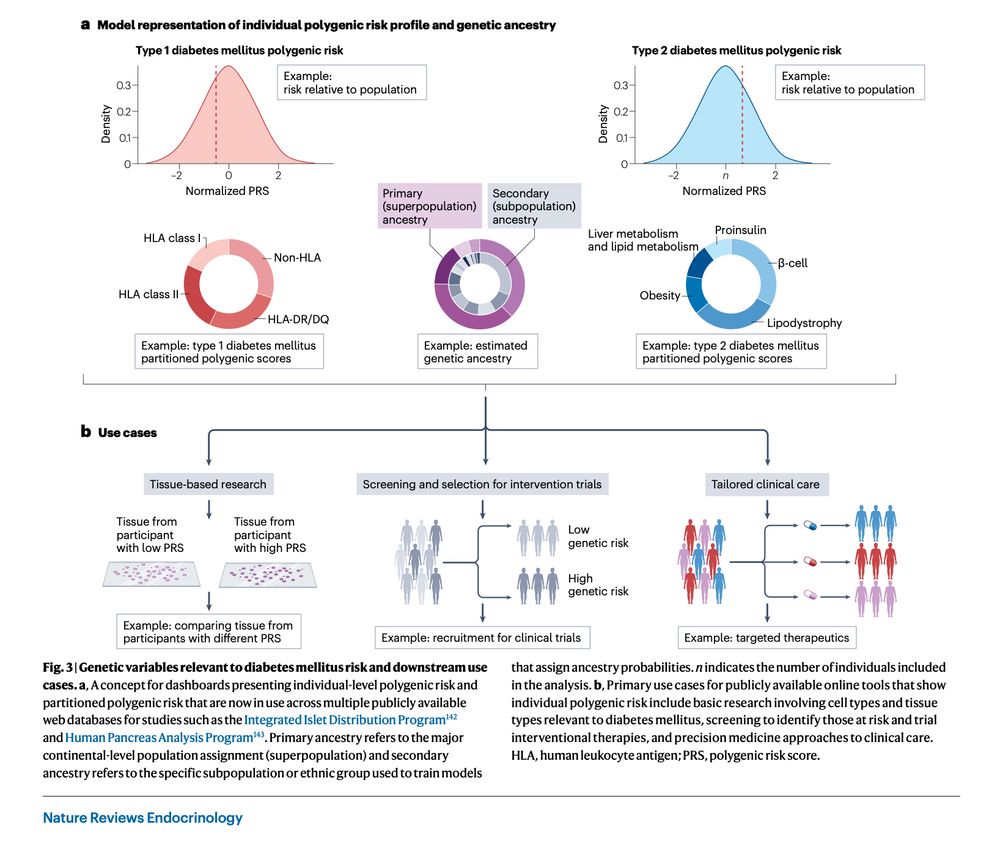
Out today from #HectorOrtega & #SethASharp A state of the art review on polygenic risk scores (#PRS) in diabetes. This is your #101 on what we currently know about them and their application to understanding disease heterogeneity & clinical translation.
Read for free here - rdcu.be/eprel

We have updated our preprint about 850,000 person UK-Danish haplotype sharing - sheding light on the rich history each person carries with them in their DNA and comparing this to historical records. Xiaolei Zhang postdoc in my group led this work. www.biorxiv.org/content/10.1...
04.06.2025 22:17 — 👍 33 🔁 10 💬 1 📌 1
We have an attractive opening for a tenure-track professorship in Single-Cell Systems Biology. Excellent package and infrastructure at @uni-wuerzburg.de Institute of Systems Immunology. Great place to live and work in the heart of the franconian wine region!
jobs.sciencecareers.org/job/673667/j...

We have 2-3 group leader positions opening @fimm-uh.bsky.social !!
We are looking for outstanding candidates in human genetics and precision medicine.
This time we have a focus on population health data science. E.g. AI for EHR/health data
Generous starting package 💰
shorturl.at/FAk6n

Nature’s careers team spoke to a dozen scientists who have been laid off from US federal agencies and from universities because of federal funding cuts in the past two months. #Academicsky 🧪
01.06.2025 01:12 — 👍 76 🔁 37 💬 1 📌 0What makes Helmholtz Munich's research unique?
At #HelmholtzMunich, our research is focused on detecting diseases earlier and preventing their progression, with the support of #AI, #genomics & #bioengineering.
👉 t1p.de/cgbwh
@www.helmholtz.de
#PersonalizedMedicine #Prevention #AI #bioengineering

Wow, so cool!🌟
Excellence in Nucleic Acid Sciences and Technologies: NUCLEATE Cluster selected for national recognition
Led by spokesperson Veit Hornung, the NUCLEATE Cluster secures funding in Germany’s Excellence Strategy
🥂🥳🤩
www.lmu.de/en/newsroom/...
@v-hornung.bsky.social