
Snippet of a chat expressing that Claude Code is a big deal
My day is summed up by this exchange with @dgmacarthur.bsky.social
25.02.2026 14:04 — 👍 8 🔁 2 💬 2 📌 0
Snippet of a chat expressing that Claude Code is a big deal
My day is summed up by this exchange with @dgmacarthur.bsky.social
25.02.2026 14:04 — 👍 8 🔁 2 💬 2 📌 0
What did the largest trial of an early detection of cancer blood test show?
erictopol.substack.com/p/the-larges...

Planning your afternoon poster session at #ashg25? Come say hello!
This is an amalgamation of our two recent preprints - working with @gregfindlay.bsky.social , @cassimons.bsky.social , @dgmacarthur.bsky.social and many others to study variation across RNU4-2 and describe a new recessive NDD 🧬
Awesome work by @zornitza.bsky.social and collaborators showing the immediate value of WGS for newborn screening in a cohort of 1,000 Australian babies. Now we need larger, more diverse cohorts to show this approach can achieve population scale!
14.10.2025 21:54 — 👍 12 🔁 6 💬 0 📌 0
🤗 Out now @naturemedicine.bsky.social results of our genomic NBS study BabyScreen+ 👶🧬
👉 www.nature.com/articles/s41...
1,000 babies
WGS using existing cards
600+ conditions
13 day TAT
16 diagnoses (vs 1 in std NBS)
High clinical impact
High parental acceptability

In these dark times, it comes as a rare pleasure to highlight @natanaels.bsky.social & @marcdemanuel.bsky.social's work on germline and somatic mutations in humans. 1/n
www.biorxiv.org/cgi/content/...
And now the fourth preprint from the TenK10K phase 1 dataset, led by @anglixue.bsky.social from @drjosephpowell.bsky.social's team - looking at genetic impacts on cell type-specific chromatin accessibility in 1,000 individuals who also have WGS and scRNA-seq!
01.09.2025 23:02 — 👍 14 🔁 0 💬 0 📌 0
RESEARCH ARTICLE • Volume 39, Issue 1001, P221- 222, November 05, 1842 A MAN WITH THREE TESTICLES. F. Macann
I think 182 year old research articles should be free
I want to read about the man
One for the reading pile and this scQTL x Disease in a MR framework feels a v powerful approach (more tissues / cell types please!) -
01.09.2025 08:17 — 👍 11 🔁 2 💬 0 📌 0Another preprint from the TenK10K program! This work, led by @alberthenry.bsky.social and Anne Senabouth, leverages the unprecedented power of this WGS/single cell RNA-seq cohort to explore causal influences of blood gene expression on immune diseases and traits. Thread:
01.09.2025 07:42 — 👍 12 🔁 2 💬 0 📌 0
Our latest research is out today on @medrxivpreprint.bsky.social:
www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n
🚨 New preprint led by amazing duo @rociorius.bsky.social and @alexblakes.bsky.social in collaboration with @cassimons.bsky.social, @dgmacarthur.bsky.social and many other amazing folks! ❤️
We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇
New preprint! The outcome of a wonderful collaboration with @nickywhiffin.bsky.social’s team to define a new recessive syndrome associated with inherited variants in RNU4-2, the non-protein-coding gene that keeps on giving.
18.08.2025 21:34 — 👍 19 🔁 4 💬 0 📌 0#HGSA2025 week kicks off with the OurDNA symposium tomorrow (with a preview of the OurDNA browser!) Register to attend online here👇
12.08.2025 23:20 — 👍 5 🔁 1 💬 0 📌 0
Image of an old building in Oxford with the heading 'postdoc opportunities' and the text 'computational approaches to improve rare disease diagnosis and treatment' and 'Big Data Institute, University of Oxford'
📣 We are recruiting! Please share!!
Are you a bioinformatician / computational scientist who wants to apply your skills to understanding regulatory biology and improving rare disease diagnosis and treatment? 🧠 💻 🧬 🩺
We have two roles available 👇
🧵 1/4

Leena Peltonen School of Human Genetics in full-swing!
@gosiatrynka.bsky.social
@dgmacarthur.bsky.social
@bpasaniuc.bsky.social
@tuuliel.bsky.social
@hilarycmartin.bsky.social
@sashagusevposts.bsky.social
@zkutalik.bsky.social
@mashaals.bsky.social
@alemedinarivera.bsky.social
This implies there is a huge literature of small microbiome association studies that is JUST NOISE. (As observed for these other fields)
27.07.2025 09:15 — 👍 25 🔁 7 💬 1 📌 0
About a hundred reasons not to use Excel.
Yes, these are politely described as 'spreadsheet' horror stories, but we all know which bit of software we're really talking about.
(HT @pgmj.bsky.social)

Peer review is not quite dead but it’s on life support and the current model of scientific publishing is a burning platform. healthydebate.ca/2025/06/topi...
26.06.2025 10:49 — 👍 37 🔁 8 💬 2 📌 0
UMAP of the 9 populations and 86 cell types identified after quality control and clustering
Preprint out today!
A team led by @tobioinformatics.bsky.social and Bradley Harris in @carlanderson.bsky.social ‘s lab has created the largest single-cell atlas of IBD tissues to date
www.medrxiv.org/content/10.1...

Hey Australian genetics/genomics friends: the OurDNA Symposium will be in Sydney on 14 August, just before the HGSA meeting. Learn more about inclusive recruitment for genomics and get a preview of the OurDNA variant browser! events.humanitix.com/ourdna-sympo...
23.06.2025 01:03 — 👍 10 🔁 9 💬 0 📌 1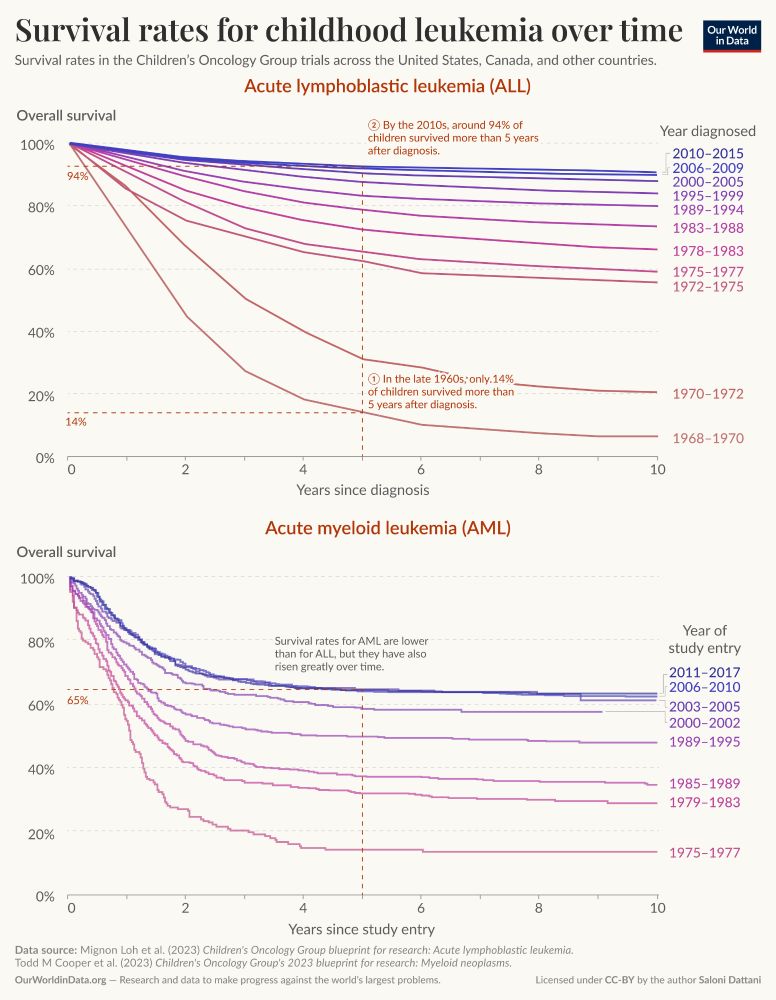
Two charts present survival rates for childhood leukemia over time, specifically focusing on Acute Lymphoblastic Leukemia (ALL) and Acute Myeloid Leukemia (AML). In the top panel, for ALL, a series of curved lines represent overall survival rates plotted against years since diagnosis. The lines show a marked increase in survival rates from the late 1960s, when only 14% of children survived more than five years post-diagnosis, to around 94% in the 2010s. Key intervals are labeled, with different colors indicating different periods of diagnosis, ranging from 1972-1975 to 2010-2015. The bottom panel illustrates survival rates for AML, which are consistently lower overall compared to ALL. Like the top graph, it features several colored lines indicating specific periods. The highest point noted indicates a survival rate of 65%. The graph captures trends in survival as well, showing gradual improvement over time, from 1975-1977 up to 2011-2017. Data sources for these visualizations are cited at the bottom: Mignon Loh et al. (2023) for ALL and Todd M Cooper et al. (2023) for AML, both from the Children's Oncology Group. The chart is published by Our World in Data, and licensed under Creative Commons by the author, Saloni Dattani.
I wrote a new piece on how much progress has been made in treating childhood leukemia.
The answer is: quite a lot!
Before the 1970s, fewer than 10% of children diagnosed survived 5 years after diagnosis.
Now most are cured and around 85% survive that long.
ourworldindata.org/childhood-le...
I worry that not enough of a big deal is being made about how long-term the devastation of these budget cuts to our scientific and health agencies will be, beyond the absolute ruin they will cause in the acute period.
03.06.2025 16:51 — 👍 2283 🔁 498 💬 47 📌 54
Flybase lost all of the NIH support overnight - it is a disaster for the community. Please consider donating. I just did! www.philanthropy.cam.ac.uk/give-to-camb...
03.06.2025 18:23 — 👍 144 🔁 148 💬 4 📌 8
Jesus. Excellent work by Michael Le Page, and utterly infuriating scenario. As Michael points out, the press release from Colossal called these dire wolves throughout. But now they want to argue that they never claimed that. Scandalous, really.
www.newscientist.com/article/2481...

Excited to be in Milan for #eshg2025! If you’re interested in leading analysis of large, diverse cohorts with WGS and cellular genomic data in Australia, I’d be keen to chat - we’re looking to fill a variety of senior comp bio roles: populationgenomics.org.au/careers/ #eshg25
24.05.2025 08:41 — 👍 14 🔁 7 💬 0 📌 0
🤗 Hugely excited to share our work on automating iterative reanalysis in #raredisease, preprint out: www.medrxiv.org/content/10.1... 🤖🧬
github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!
The Cambridge Companion to Workday
The Routledge Handbook of Dual-Factor Authentication
The Oxford Handbook of Figuring Out How To Add A Signature To This PDF, Why The Hell Is This So Difficult

A graph showing the price per pound for different vehicles and types of cheese. There are a surprising number of cheeses in the top half, such as Roquefort.
This is the best graph I've seen in a while 📊
17.05.2025 21:30 — 👍 353 🔁 121 💬 21 📌 22