The unification of representation learning and generative modelling
A deep dive into the convergence of discriminative and generative AI, covering 4 phases of evolution from REPA to RAE and beyond.
Too many REPA / RAE / representation alignment papers lately?
I was lost too, so I wrote a blog post that organizes the space into phases and zooms in on what actually matters for general/molecular ML.
Curious what folks think - link below!
🔗 Blog: kdidi.netlify.app/blog/ml/2025...
12.02.2026 06:27 — 👍 11 🔁 2 💬 0 📌 0

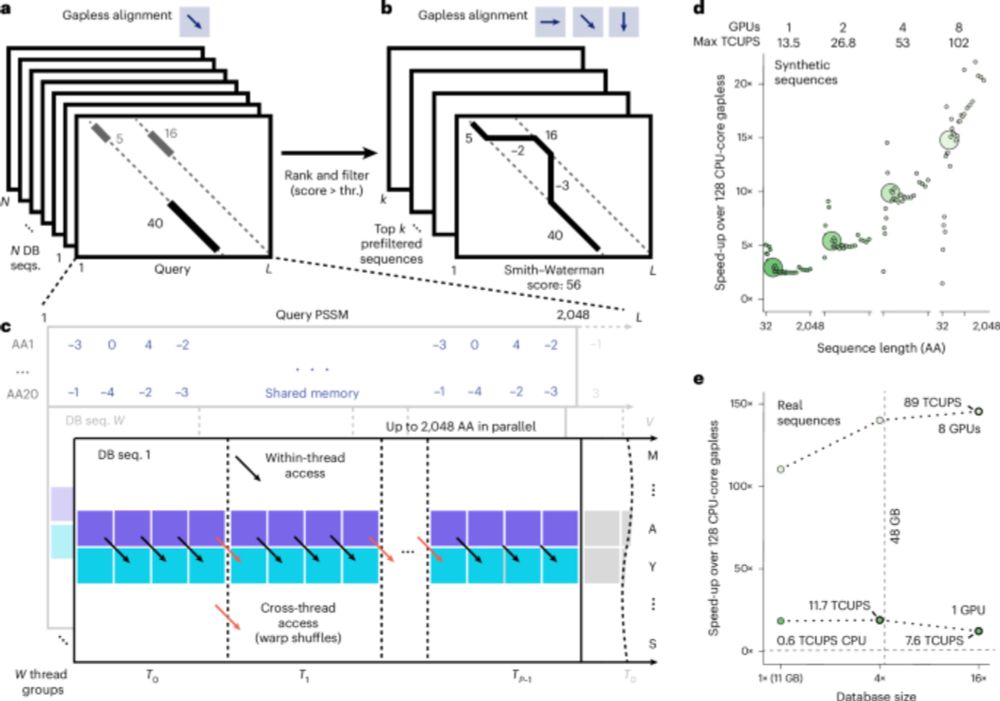
GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
21.09.2025 08:06 — 👍 174 🔁 63 💬 4 📌 2
For more details read the thread by the man himself: bsky.app/profile/ncor...
15.08.2025 17:49 — 👍 1 🔁 0 💬 0 📌 0
An incredible project to witness, led by the most incredible dreamteam @ncorley.bsky.social , @simonmathis.bsky.social and Rohith Krishna with an amazing team inside and outside the Baker lab. Check it out and let us know what you think/contribute to the codebase! 6/6
15.08.2025 17:48 — 👍 1 🔁 0 💬 1 📌 0
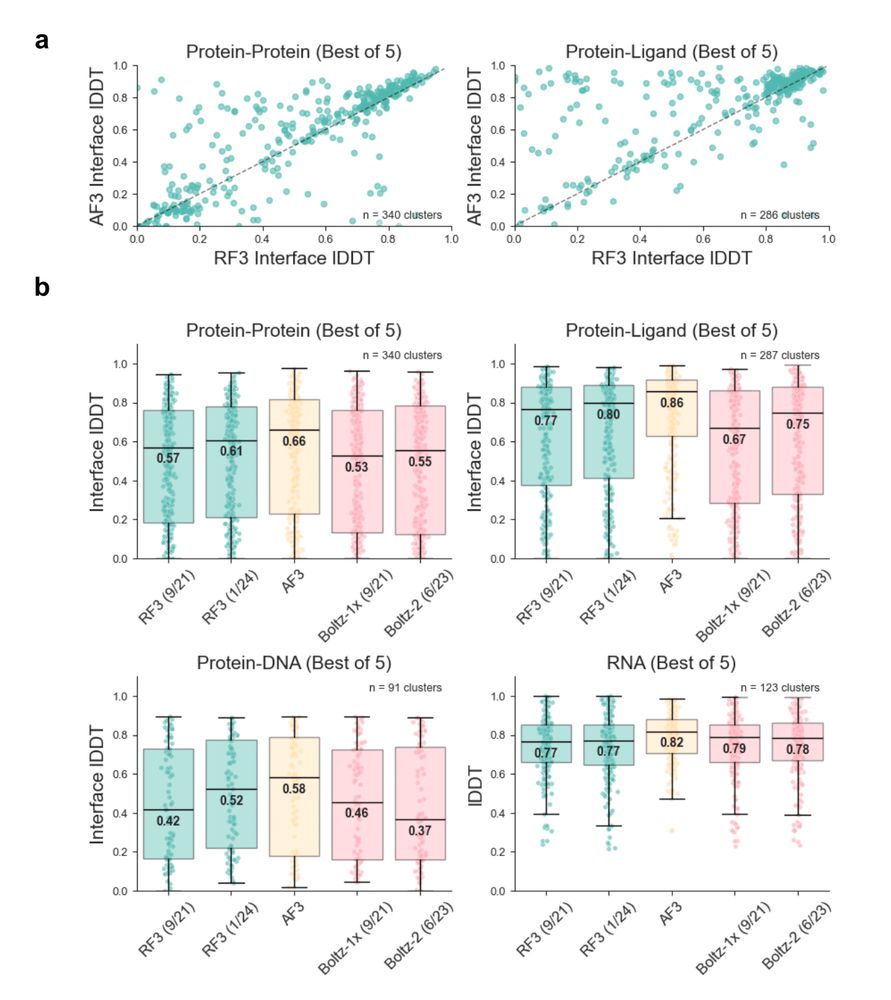
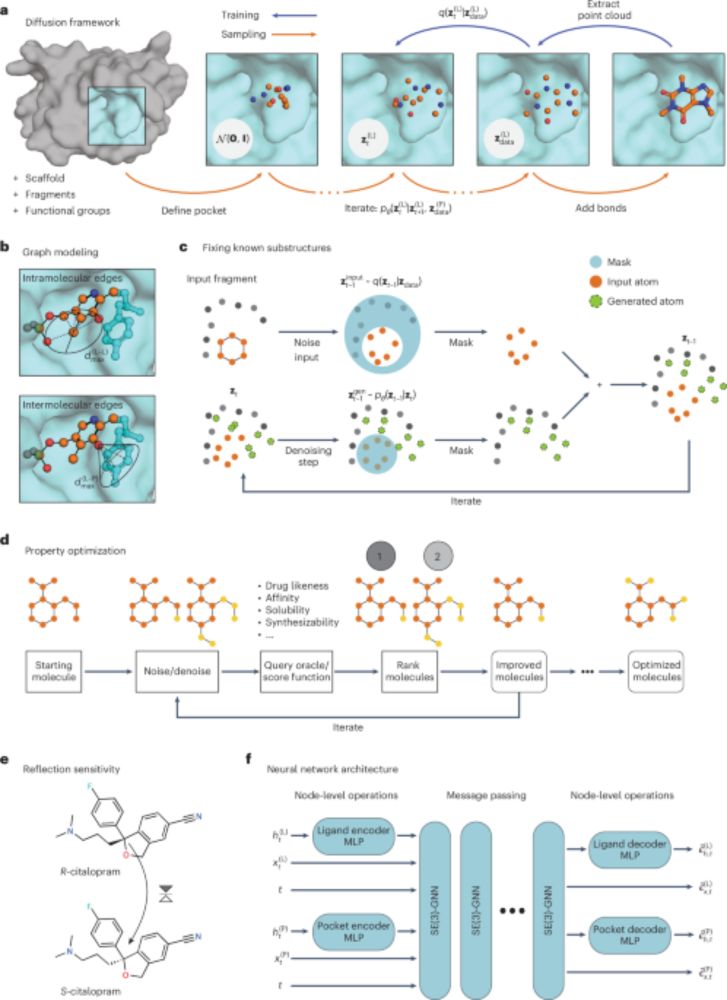
The preprint shows how atomworks leads to better reference conformers (and better predictions!), enables advanced features in RF3 like chirality-aware training or ligand templating and narrows the performance gap to closed-source models. 5/6
15.08.2025 17:47 — 👍 0 🔁 0 💬 1 📌 0

`atomworks.ml` on the other hand offers advanced dataset featurization and sampling for deep learning workflows, all operating on the canonical AtomArray object from @biotite_python so that all transforms are traceable and generalizable between models. 4/6
15.08.2025 17:46 — 👍 2 🔁 1 💬 1 📌 0

AtomWorks has two main components: atomworks.io takes a file (cif, sdf, ...) and does parsing, cleaning and more. You can also look at your structures in a notebook or via PyMol thanks to pymol-remote, so you can directly inspect if your code does what you want! 3/6
15.08.2025 17:45 — 👍 1 🔁 1 💬 1 📌 0

In the past, every BioML model had its own data pipeline, creating loads of overhead. With AtomWorks, >80% of code is shared between models like ProteinMPNN, RF3 or design models. 2/6
15.08.2025 17:44 — 👍 0 🔁 0 💬 1 📌 0

AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
15.08.2025 17:44 — 👍 22 🔁 6 💬 1 📌 0

RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
15.08.2025 13:26 — 👍 54 🔁 19 💬 0 📌 0
Very excited about our latest all-atom generative model proteina, check out the project page (research.nvidia.com/labs/genair/...) and stay tuned for the code release soon!
19.07.2025 20:46 — 👍 24 🔁 9 💬 0 📌 1
Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR team at NVIDIA.
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
23.04.2025 08:37 — 👍 12 🔁 2 💬 0 📌 0
Such a fun project to work on with a stellar team! Stay tuned for other things to come here, and see you all in Singapore!
04.03.2025 17:44 — 👍 5 🔁 1 💬 0 📌 0
Yet another story of issues with benchmarks and evaluations in ML4bio + a much stronger and fair benchmark #bioMLeval
20.02.2025 06:00 — 👍 12 🔁 2 💬 0 📌 0
Have a look at our shiny new benchmark for motif-scaffolding in computational protein design! New (and harder) tasks, including a reproducible evaluation pipeline
20.02.2025 15:51 — 👍 2 🔁 0 💬 0 📌 0
This!
Also well put in this editorial in PLOS Comp Biol:
Putting benchmarks in their rightful place: The heart of computational biology
doi.org/10.1371/jour...
15.01.2025 12:09 — 👍 19 🔁 6 💬 0 📌 0
Our paper on computational design of chemically induced protein interactions is out in @natureportfolio.bsky.social. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia!
t.co/vtYlhi8aQm
15.01.2025 16:37 — 👍 65 🔁 25 💬 1 📌 0
So excited for you and what is to come! Onwards and Upwards;)
23.12.2024 22:44 — 👍 2 🔁 0 💬 0 📌 0
Thanks Jascha 🫶
We’re working hard to create sustainable funding mechanisms for open source scientific tooling - understanding the challenge landscape is a key first step!
03.12.2024 10:45 — 👍 9 🔁 2 💬 0 📌 0
Love PyMOL Remote, one of these tools that does one thing and does it well!
29.11.2024 11:28 — 👍 7 🔁 1 💬 0 📌 0
This should go chiral
27.11.2024 20:35 — 👍 149 🔁 17 💬 7 📌 0
MSAs go brrr with MMseqs2-GPU! Super fun project, happy to work with and learn from a stellar team of engineers and scientists. Try it out and stay tuned!
📄 Preprint: www.biorxiv.org/content/10.1...
💾 Code: mmseqs.com
🗞️ Blog: developer.nvidia.com/blog/boost-a...
16.11.2024 14:18 — 👍 29 🔁 7 💬 0 📌 1
AI researcher excited about biomolecule design 🧬
PhD student at the University of Cambridge
Prev. at FAIR, Prescient Design, and MRC LMB
📝 https://chaitjo.substack.com
PhD Candidate @ the University of Washington's Institute for Protein Design | Baker Lab | Machine Learning for Protein Design | Enzymes
Passionate for integrated structural biology, including NMR and many more. Particularly interested in protein dynamics, and therefore chaperones, enzymes and mitochondrial protein import.
Fortunate to lead a great research team at IST Austria.
Supramolecular Chemist (PI @ Durham University) interested in enzyme mechanism and mimicry, cages and catalysis
This Python package is your Swiss army knife for bioinformatics. It allows easy and fast analysis of sequence and biomolecular structure data.
Assistant professor of chemistry at the Technical University of Denmark (DTU). Also at Jura Bio. machine learning, statistics, chemistry, biophysics
https://eweinstein.github.io/
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
PhD student @sangerinstitute
Schmidt Science Fellow | Postdoc @ Stanford | Prev. DPhil @ Oxford || AI for Molecule & Cell Modeling
Doctoral researcher at the University of Oxford. Interested in social and political philosophy, metaphilosophical questions and feminism. she/her
Principal Research Scientist at NVIDIA | Former Physicist | Deep Generative Learning | https://karstenkreis.github.io/
Opinions are my own.
Neuronerd, Prof, head of HIP lab in Experimental Psychology, Oxford. Researcher, UK AI Safety Institute. https://humaninformationprocessing.com/.
Assistant Professor at Stanford Statistics and Stanford Data Science | Previously postdoc at UW Institute for Protein Design and Columbia. PhD from MIT.
Not-for-profit Focused Research Organisation (FRO) turning disordered proteins into viable drug targets | London, UK | https://bindresearch.org/
Creating enzymes by computational design and directed evolution.
Group Leader at MPI for Terrestrial Microbiology, Germany
https://www.mpi-marburg.mpg.de/home
#FascinatedByEnzymes
Interested in small molecule drug design.
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
PhD student at EPFL working on generative molecular design | Previously Microsoft AI4Science and AstraZeneca