Paper: authors.elsevier.com/a/1lWEe8YyDf...
29.07.2025 19:04 — 👍 1 🔁 0 💬 0 📌 0Possu Huang Lab
@possuhuanglab.bsky.social
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
@possuhuanglab.bsky.social
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
Paper: authors.elsevier.com/a/1lWEe8YyDf...
29.07.2025 19:04 — 👍 1 🔁 0 💬 0 📌 0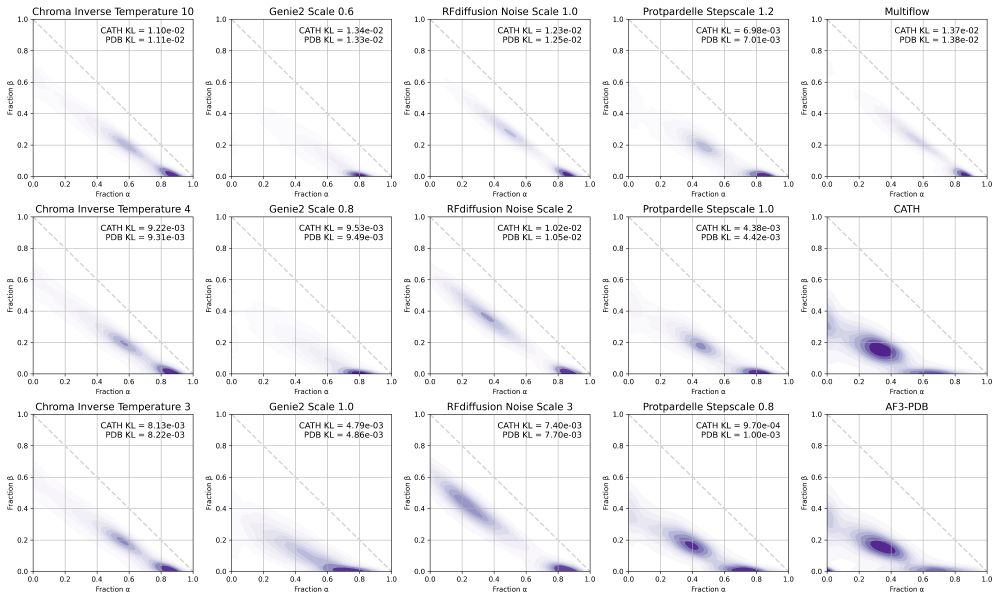
We include some additional analysis in the supplement, including secondary structure distributions.
29.07.2025 19:04 — 👍 0 🔁 0 💬 1 📌 0SHAPES now published in Cell Systems!
29.07.2025 19:04 — 👍 2 🔁 0 💬 1 📌 0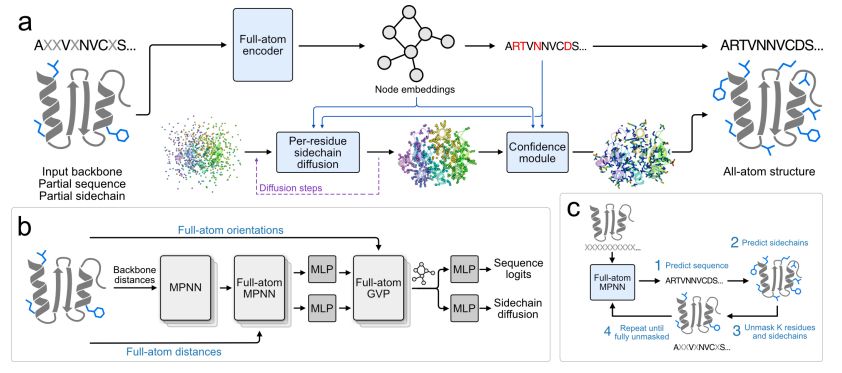
FAMPNN architecture
All-atom fixed backbone protein sequence design with FAMPNN
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
I'm organizing a Keystone symposium, along with Liz Kellogg and @possuhuanglab.bsky.social, on machine learning and macromolecules. Mar 23-26 in Keystone, Colorado. We have a great lineup and deadlines are coming up soon!
15.01.2025 20:47 — 👍 45 🔁 18 💬 0 📌 1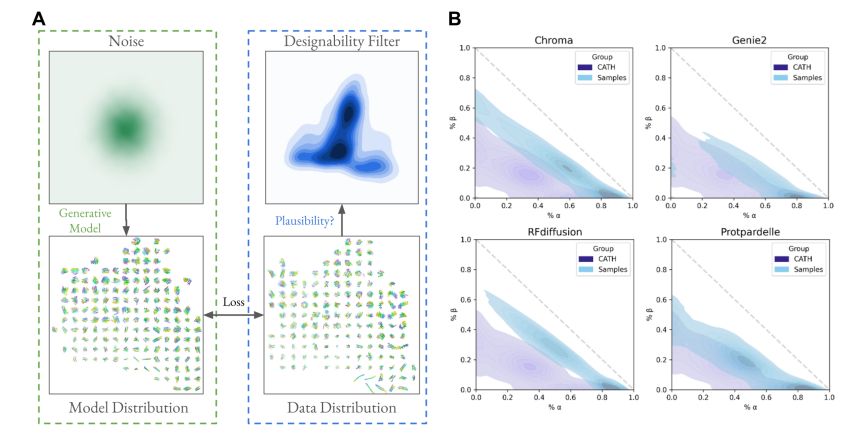
Generative models capture a biased set of protein structure space
Generative models do not capture the full expressivity of PDB structures
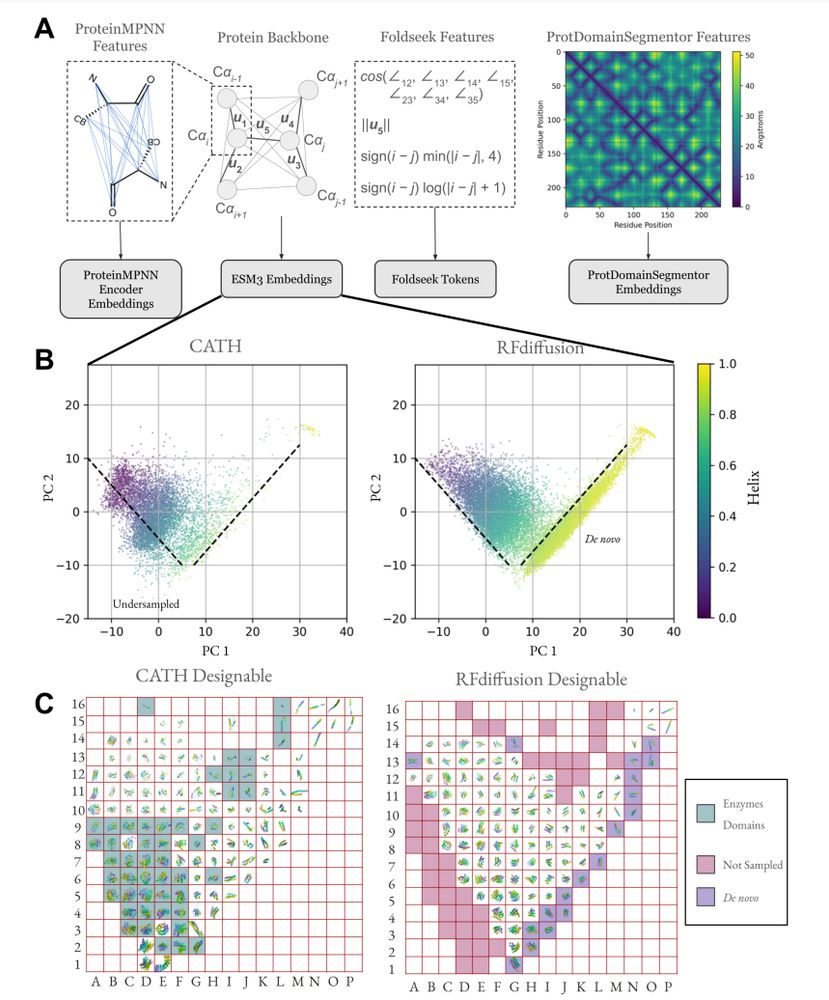
Protein structure embeddings reveal undersampled and de novo structure space
A framework for evaluating how well generative models of protein structure match the distribution of natural structures.
@possuhuanglab.bsky.social
www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
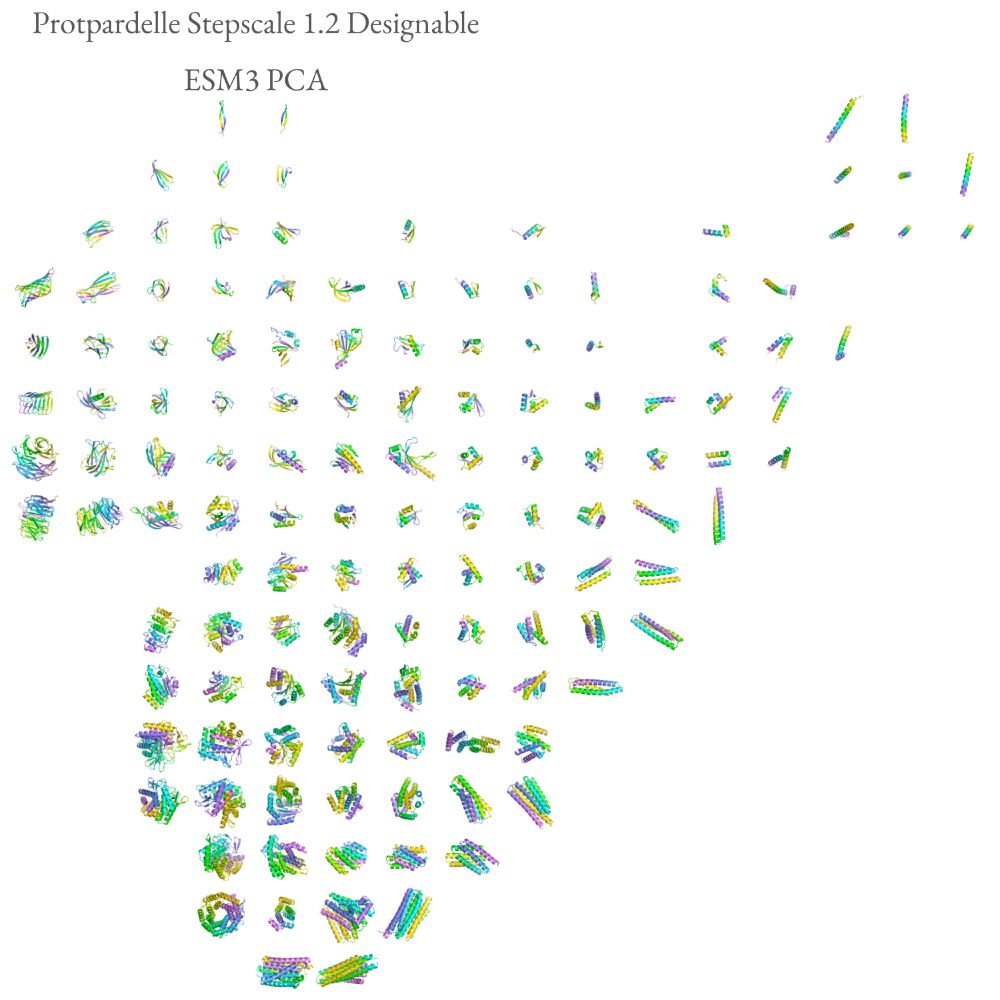
Our supplement has many additional figures of the rasterized protein structure space, stratified by designable and not designable and spatially organized by ESM3 and ProtDomainSegmentor embeddings.
15.01.2025 18:48 — 👍 1 🔁 0 💬 1 📌 0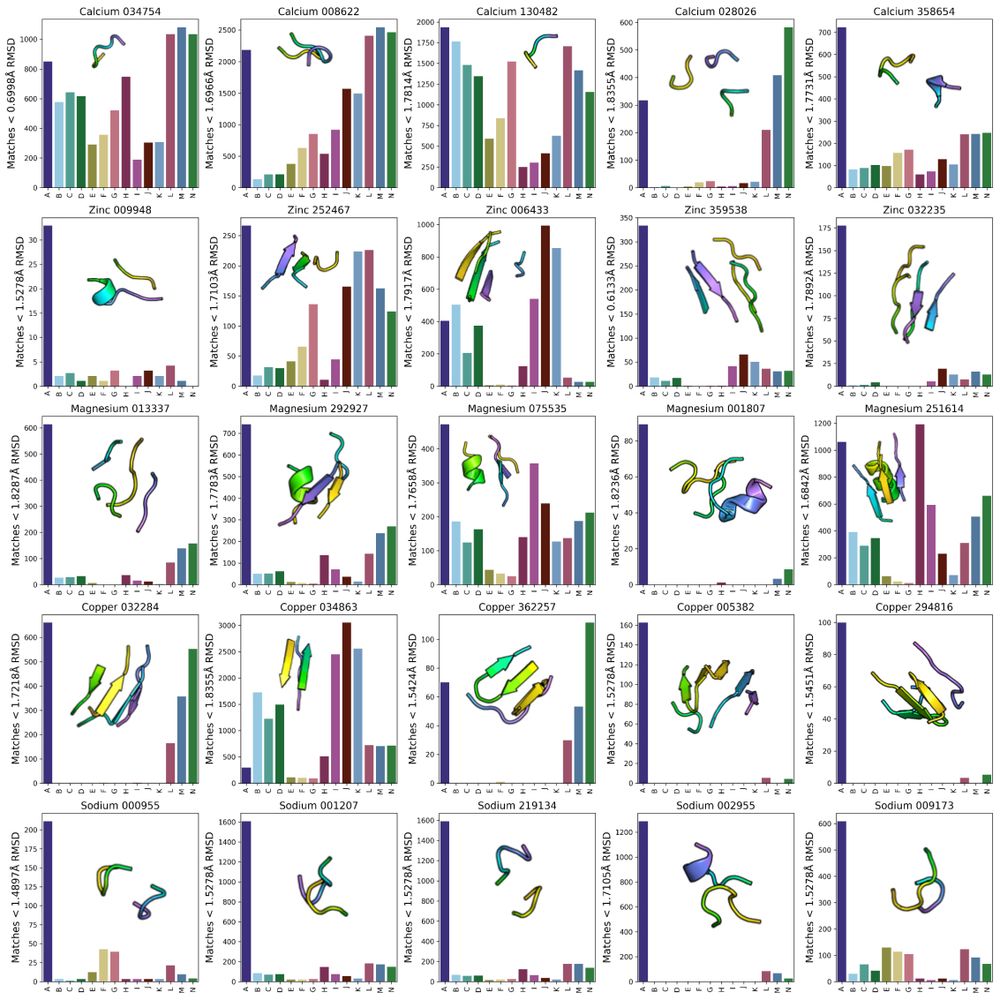
One consequence of unbiased sampling of protein structure space is a higher likelihood of finding TERtiary Motifs (TERMs) which involve complex loops, with implications for functional protein design (see Figure 5 legend for group labels).
15.01.2025 18:48 — 👍 1 🔁 0 💬 1 📌 0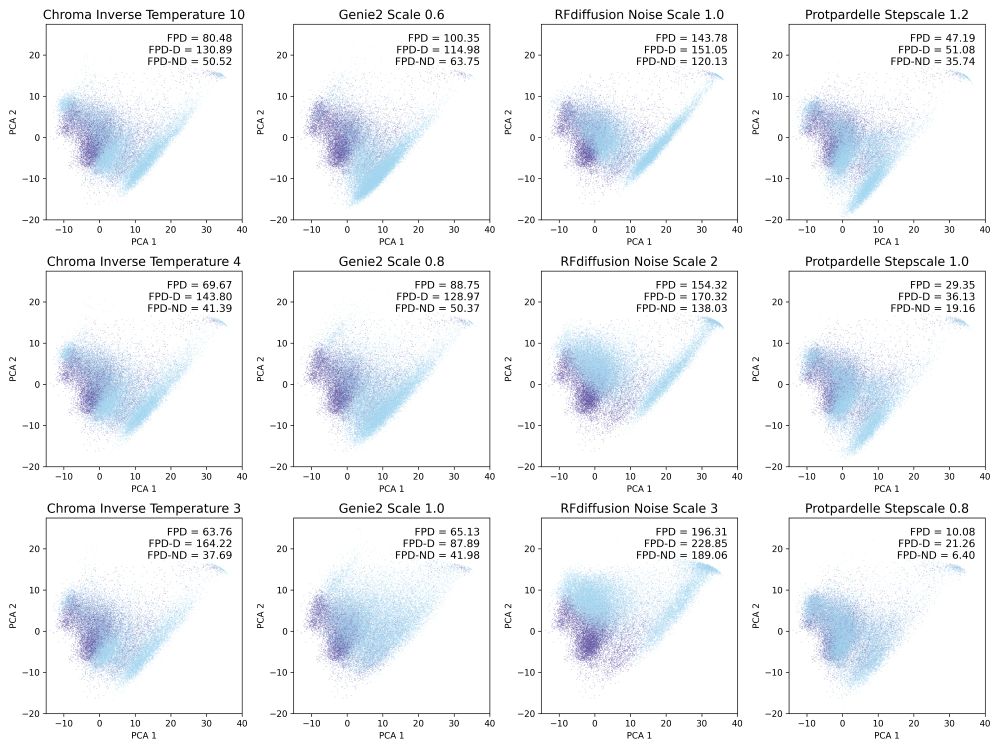
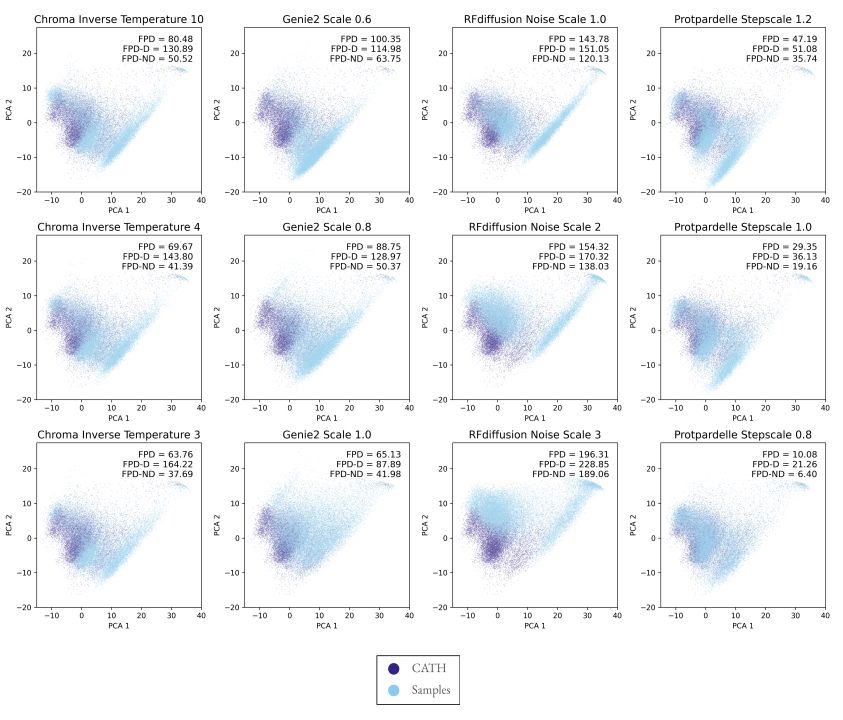
Inspired by the FPD metric in EvoDiff for protein sequence distributions, we compute Fréchet distance using protein structure embeddings, also subsetted to designable and non-designable samples (FPD-D and FPD-ND).
15.01.2025 18:48 — 👍 0 🔁 0 💬 1 📌 0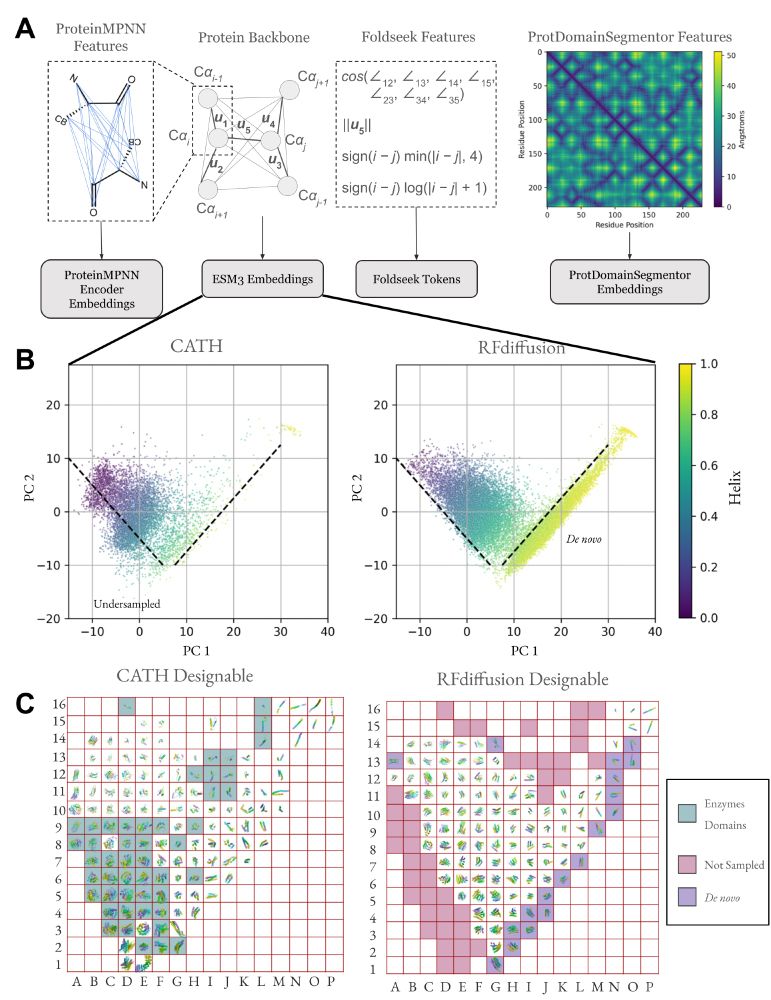
New preprint from our group! We propose SHAPES, a set of metrics to quantify the distributional coverage of generative models of protein structures with embeddings at different structural hierarchies and quantify undersampling / extrapolation behaviors.
15.01.2025 18:48 — 👍 28 🔁 7 💬 1 📌 1This is a clever way to use synthetic biology: taking a toxin that overactivate the immune system (superantigen), rationally modify its core components and transform it into a platform of immunotherapy agents.
Congratulations to @haotiandu.bsky.social and @possuhuanglab.bsky.social on the 2 papers!
Checkout out these two bombshell papers from @possuhuanglab.bsky.social @stanfordmedicine.bsky.social, computational design of antigen-specific binders to MHC-I or -II, with applications to next gen targeted therapeutics 🤯
17.12.2024 07:16 — 👍 89 🔁 22 💬 0 📌 1Science in 60 Seconds: Haotian Du, PhD student with Possu Huang’s lab, explains her research on creating novel proteins that expands the possibilities for detecting more cancer types.
@possuhuanglab.bsky.social @haotiandu.bsky.social
Incredible work from BioE Professor’s Possu Huang Lab @possuhuanglab.bsky.social @haotiandu.bsky.social - pioneering novel proteins with new structures and functions, unlocking new possibilities for detecting more cancer types. Congratulations!
17.12.2024 01:51 — 👍 5 🔁 1 💬 0 📌 0@haotiandu.bsky.social is now on BlueSky 😄
17.12.2024 01:33 — 👍 2 🔁 0 💬 0 📌 0Excited to share that TRACeR stories are out🙌
17.12.2024 01:12 — 👍 5 🔁 3 💬 0 📌 0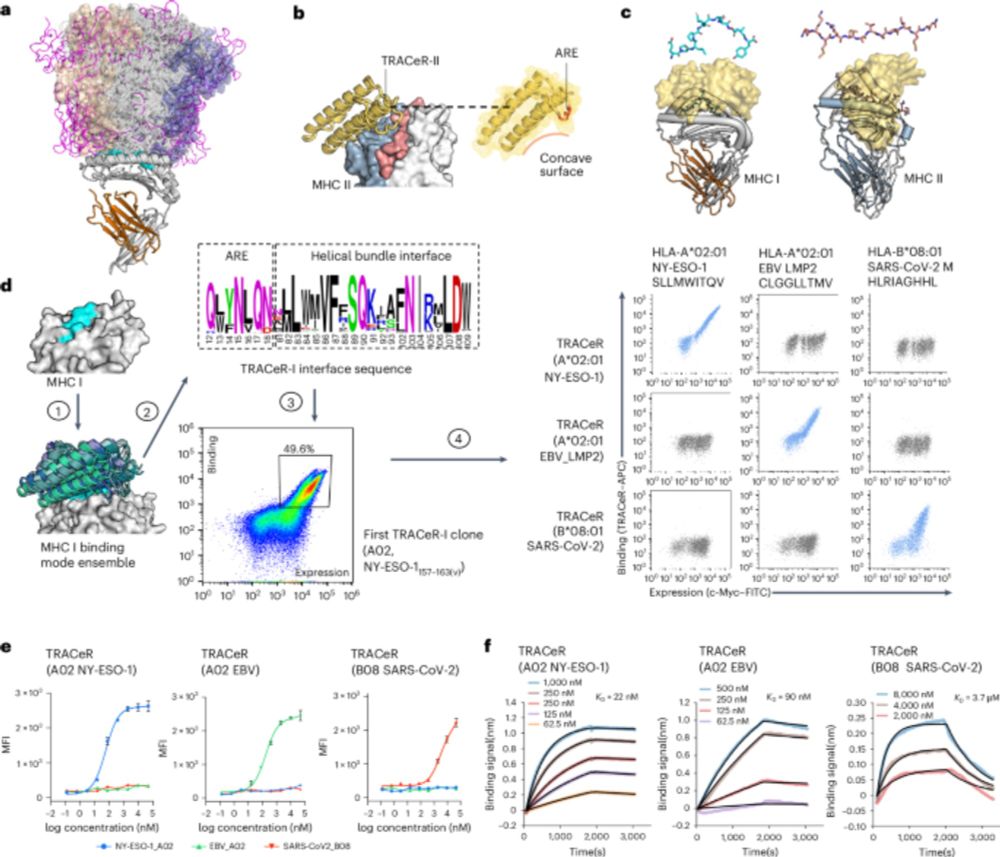
A new system, TRACeR-I, targets peptides on MHC I molecules with high precision, enabling specific tumor and virus cell killing.
It powers CAR-T and BiTE therapies, showing potential to transform cancer and infectious disease treatment.
www.nature.com/articles/s41...
The TRACeR project was led by Haotian Du and Jingjia Liu and a collaborative effort between the Huang Lab at @bioe-stanford.bsky.social, Stanford BioX and the Sgourakis Lab at UPenn/CHOP
@naturebiotech.bsky.social @stanfordmedicine.bsky.social @uofpenn.bsky.social @stanfordbiosci.bsky.social

7/ Our platform offers a convenient strategy for creating MHC binders: from a master library, peptide-specific MHC binders can be isolated simply by a few rounds of yeast surface display enrichment.
17.12.2024 00:56 — 👍 2 🔁 0 💬 1 📌 0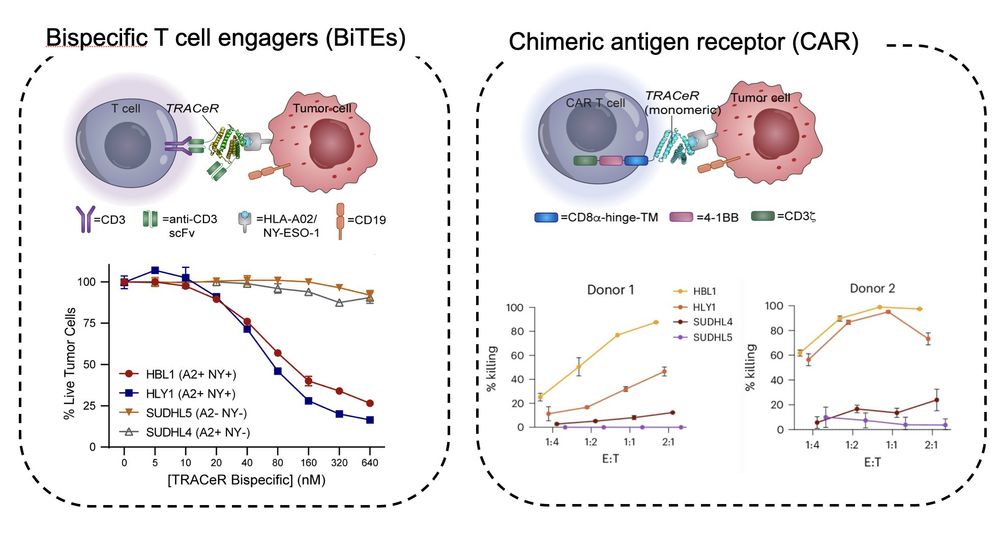
6/ We show proof-of-concept targeted killing of tumor cells using TRACeR-I.
17.12.2024 00:56 — 👍 1 🔁 0 💬 1 📌 0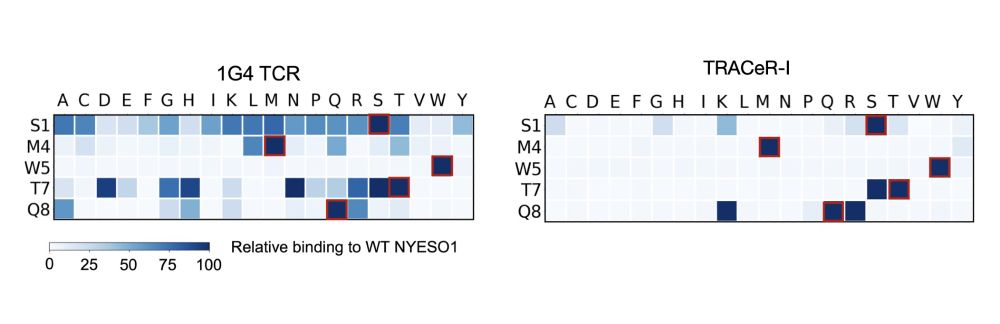
5/ The mechanism of TRACeR-1 can achieve discrimination of single point mutants to peptides.
17.12.2024 00:56 — 👍 1 🔁 0 💬 1 📌 0
4/ But MHC-I peptides have bulged conformations. For TRACeR-I, we use a set of spatially-defined amino acid residues to “box-in” on the peptide and achieve recognition.
17.12.2024 00:56 — 👍 1 🔁 0 💬 1 📌 0
3/ Peptides in MHC-II are in an extended conformation. TRACeR-II uses a surface loop to reach into MHC-II’s peptide binding groove to recognize the peptide. We use a single loop to convey specificity to the peptide in the binding groove.
17.12.2024 00:56 — 👍 2 🔁 0 💬 1 📌 0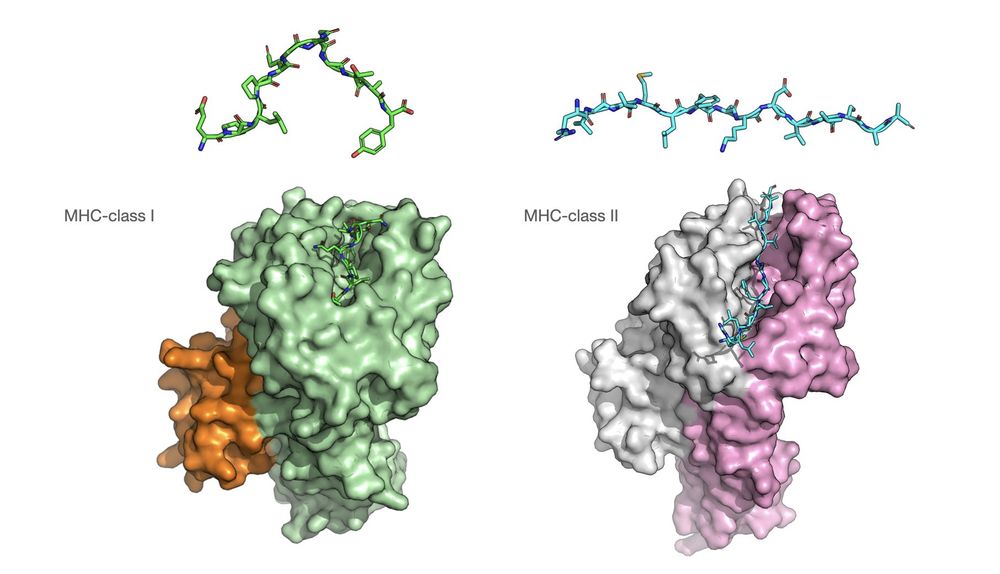
2/ MHC antigens are central to the immune system and can be leveraged for disease-specific targeting. These peptides are derived from intra-cellular proteins and ingested foreign antigens for presentation by class-I and class-II MHCs, respectively.
17.12.2024 00:56 — 👍 0 🔁 0 💬 1 📌 0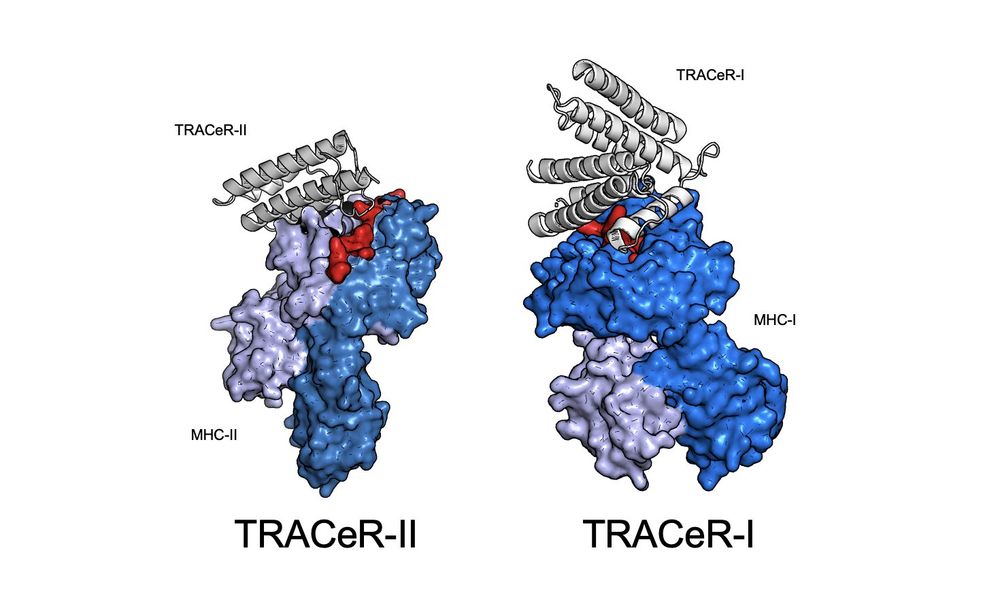
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk