Interested in a co-authorship?
We’re building a tool for repository-scale untargeted #metabolomics and #exposomics of #environmental data. To make it the best it can be, we’re looking for people willing to share high-resolution LC-MS/MS (DDA) data from #water, #soil, #sediment, and related samples.
26.08.2025 19:51 — 👍 16 🔁 13 💬 3 📌 1
Adobe Acrobat
📢 Join the EMN Committee! Applications NOW OPEN—we want you on our 2025‑26 team. Make a difference, grow your skills, and connect with peers.
🗓️ Jul 28–Aug 10 2025
🔗 Info: tinyurl.com/mr2zkhhc | ✍️ Apply: shorturl.at/0uams
#EMNCommittee #ECR #MetabolomicsSociety
28.07.2025 06:19 — 👍 4 🔁 4 💬 0 📌 1
There is still time to apply for #PhD studentships with us!
Thanks for spreading the word.
#phdchat #phdlife #academiclife
19.05.2025 19:59 — 👍 5 🔁 7 💬 0 📌 0
As the manuscript is currently under review, I welcome any feedback or question 😁
06.05.2025 14:57 — 👍 0 🔁 0 💬 0 📌 0
This project was a massive collaborative effort between @dukepress.bsky.social, @caltech.edu, and @ucsandiego.bsky.social. A big thank you goes to the PIs who made this possible - Rima Kaddurah-Daouk, Sarkis Mazmanian, Rob Knight, and @pieterdorrestein.bsky.social - and all the collaborators! 13/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 1
We believe this resource and its associated tools will aid future research in the pathophysiology of AD and in the broader context of #biomedicalresearch. Additionally, this dataset contains hundreds of yet-to-be-characterized metabolites that researchers can explore. 12/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
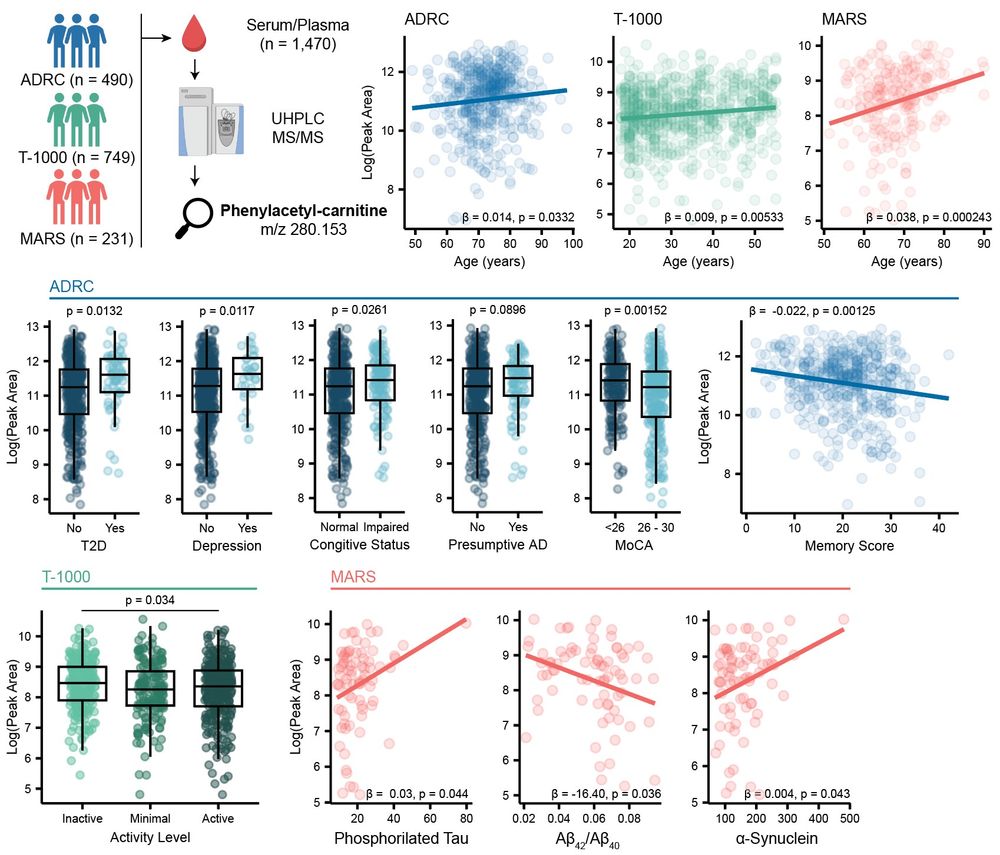
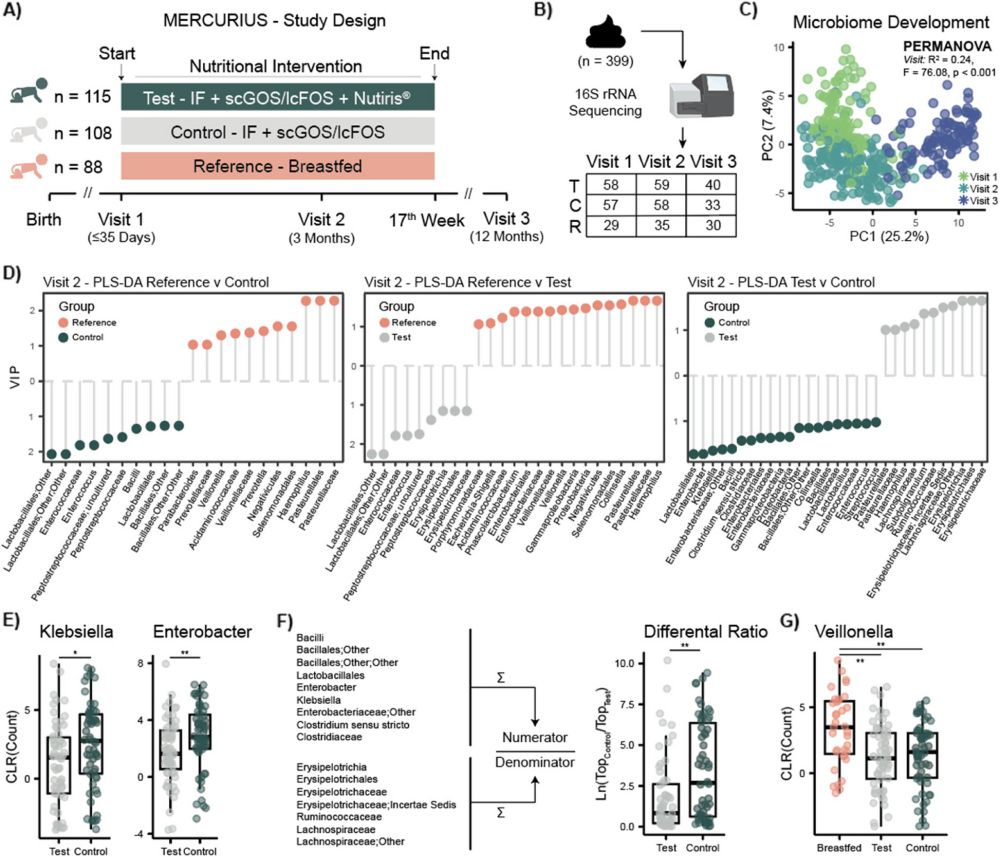
We then reprocessed 1.5k blood samples collected by the Alzheimer Gut Microbiome Project (AGMP) and found phenylacetyl-carnitine to be correlated with #aging, #cognitive impairment, #physical exercise and AD #biomarkers in the cerebral spinal fluid (CSF). 11/n
06.05.2025 14:57 — 👍 0 🔁 0 💬 1 📌 0
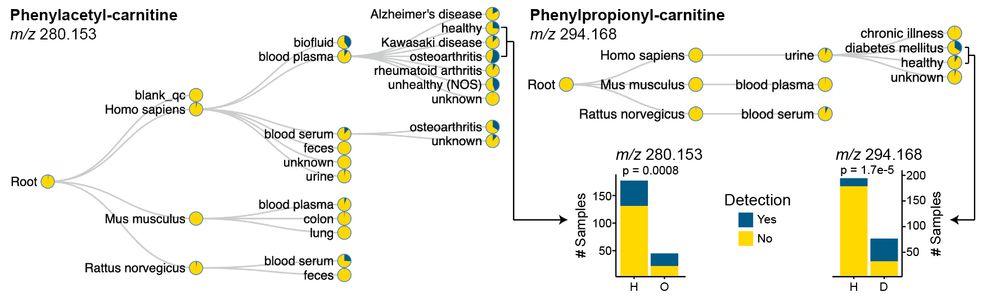
Looking for these previously #uncharacterized carnitines, we observed phenylacetyl-carnitine to be enriched in inflammatory diseases. This workflow can be applied to any kind of (un)annotated MS/MS spectrum researchers are interested in. 10/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
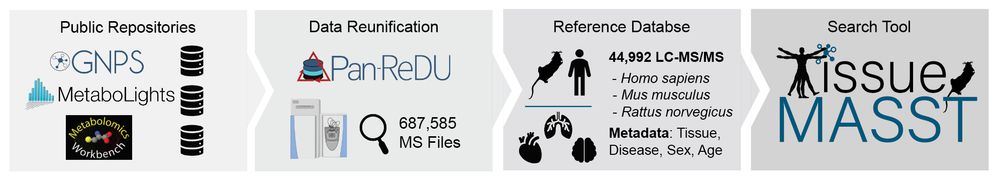
We then developed #tissueMASST to translate animal findings to human datasets. This MS/MS search tool comprises ~50k LC-MS/MS samples captured from animal models and humans with associated metadata, such as tissue type, disease status, sex, and age. 9/n
06.05.2025 14:57 — 👍 0 🔁 0 💬 1 📌 1
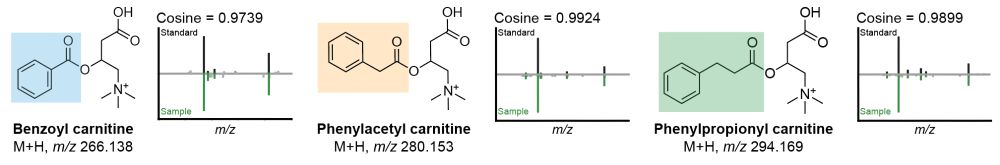
Given their possible biological relevance, we did some magic 🪄 and annotated them as benzoyl-carnitine, phenylacetyl-carnitine, and phenylpropionyl-carnitine. We #synthesized the standards and their MS/MS spectra are now part of the GNPS libraries. 8/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
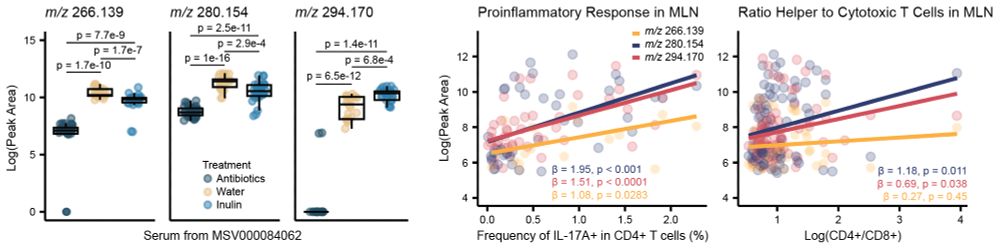
These molecules were classified as modulated by the microbiome and, in an external dataset, were observed responding both to #antibiotics and #prebiotics. Additionally, they positively correlated with #inflammation markers of the mesenteric lymph nodes of the mice. 7/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
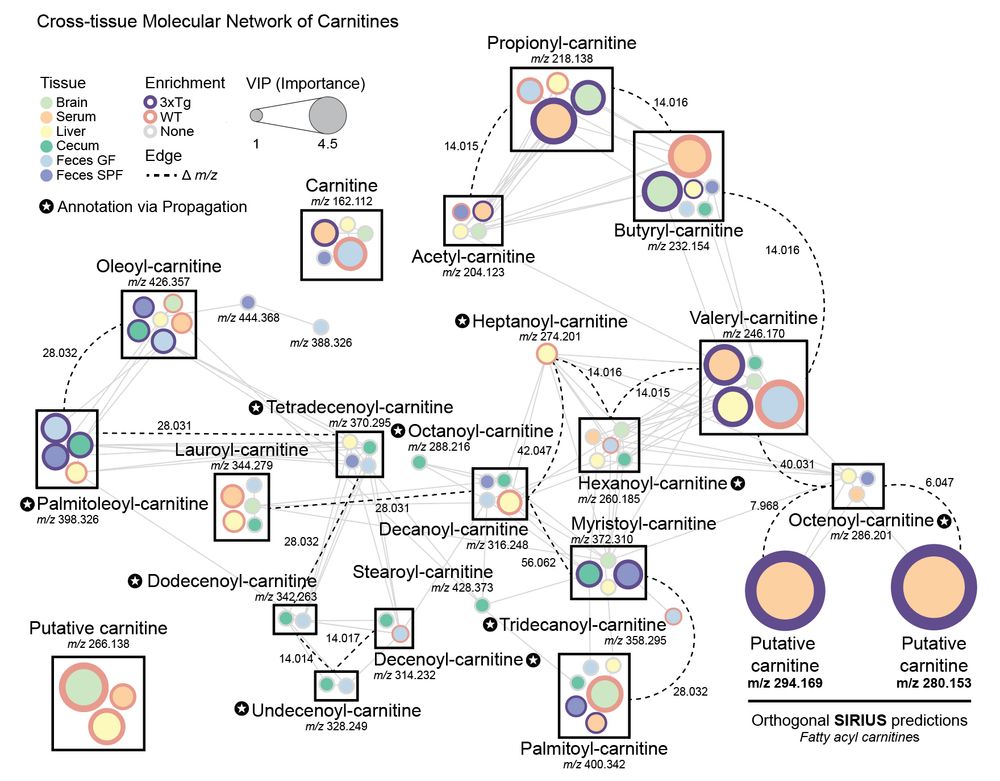
Specifically focusing on #carnitines, which were dysregulated across multiple tissues, we generated a cross-tissue molecular network to capture a holistic view. We observed 3 putative carnitines that were not annotated via GNPS MS/MS libraries. 6/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
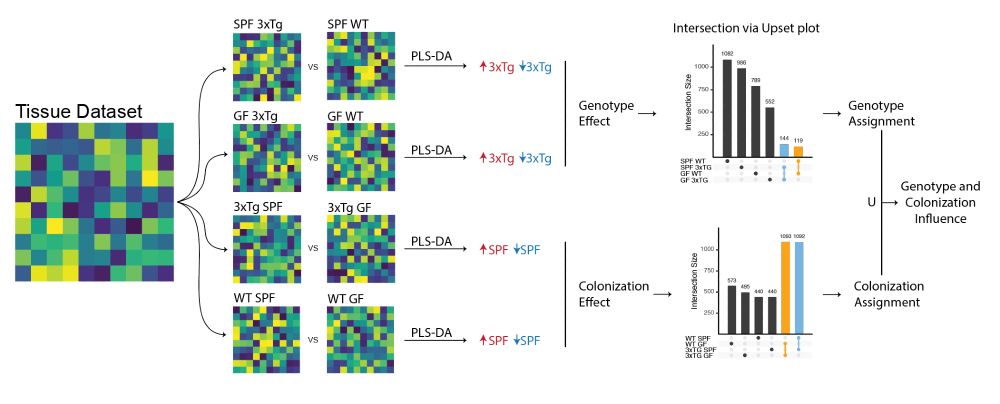
Having both SPF and GF animals also allowed us to putatively identify if molecular alterations were driven by the #genotype, the #microbiome, or both. Information available for all the molecular features differentiating AD animals from WT. 5/n
06.05.2025 14:57 — 👍 2 🔁 0 💬 1 📌 0
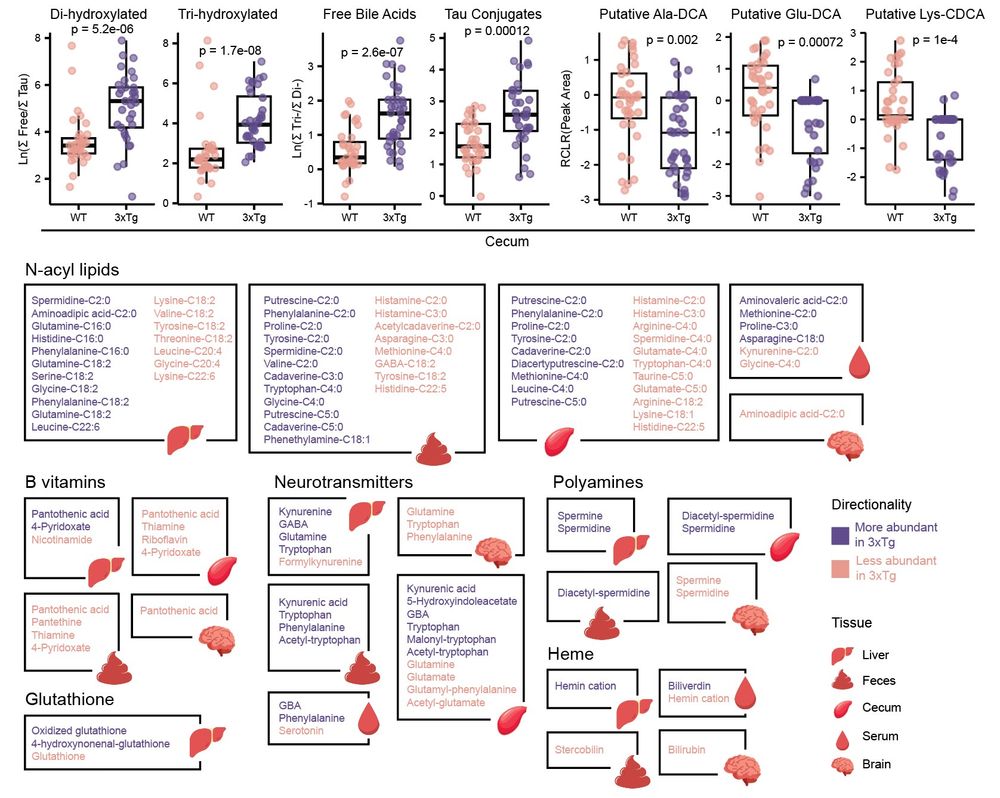
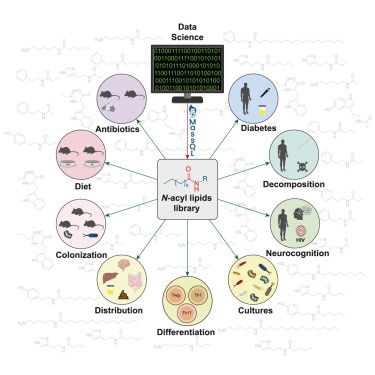
Thousands of metabolic features were driving these separations across tissues. Alterations were observed in the #bileacids pool (obviously😉) but also in N-acyl lipids, B vitamins, neurotransmitters, polyamines, and several other classes of molecules. 4/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
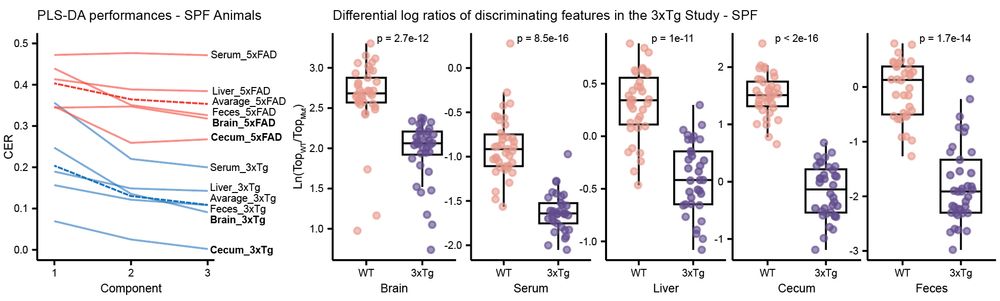
Murine models of #Alzheimers Disease (AD) present distinct metabolic profiles across five tissue types captured by this atlas - brain, serum, liver, cecum, and feces. 3/n
06.05.2025 14:57 — 👍 0 🔁 0 💬 1 📌 0
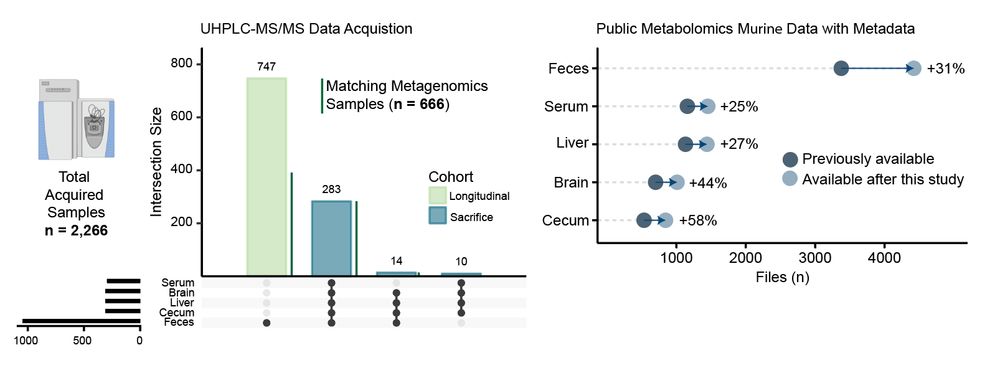
We have generated an untargeted #metabolomics atlas from more than 2,000 samples collected from tissues of 300 mice with different genetic backgrounds (3xTg, 5xFAD, C57BL/6J), colonization conditions (SPF, GF), sex, and age. Data and metadata are publicly available. 😁 2/n
06.05.2025 14:57 — 👍 1 🔁 0 💬 1 📌 0
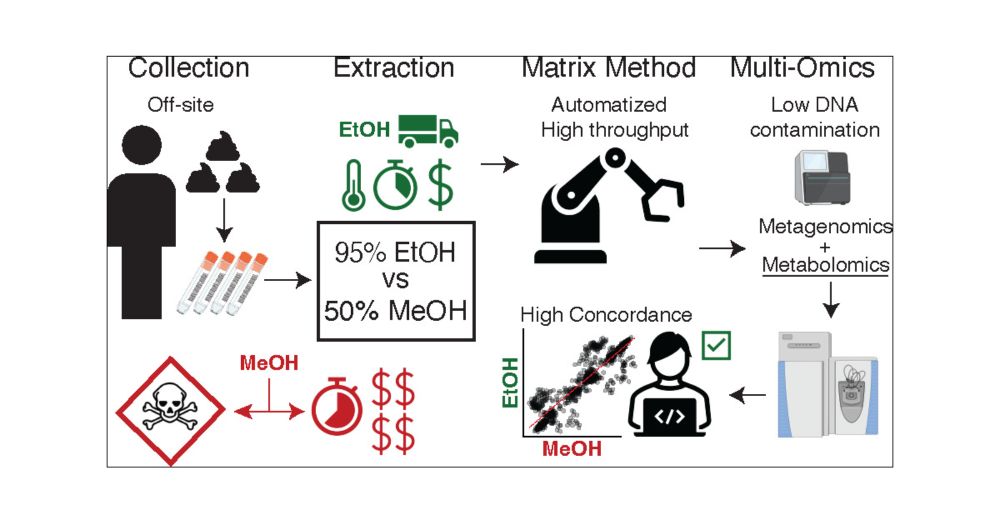
Human Untargeted Metabolomics in High-Throughput Gut Microbiome Research: Ethanol vs Methanol
Untargeted metabolomics is frequently performed on human fecal samples in conjunction with sequencing to unravel the gut microbiome functionality. As sample collection efforts are rapidly expanding, with individuals often collecting specimens at home, metabolomics experiments should adapt to accommodate the safety and needs of bulk off-site collections and improve high throughput. Here, we show that a 95% ethanol, safe to be shipped and handled, extraction part of the Matrix Method pipeline recovers comparable amounts of metabolites as a validated 50% methanol extraction, preserving metabolic profile differences between investigated subjects. Additionally, we show that the fecal metabolome remains relatively stable when stored in 95% ethanol for up to 1 week at room temperature. Finally, we suggest a metabolomics data analysis workflow based on robust centered log ratio transformation, which removes the variance introduced by possible different sample weights and concentrations, allowing for reliable and integration-ready untargeted metabolomics experiments in gut microbiome research.
Human Untargeted Metabolomics in High-Throughput Gut Microbiome Research: Ethanol vs Methanol | Analytical Chemistry pubs.acs.org/doi/10.1021/...
03.03.2025 21:17 — 👍 19 🔁 7 💬 0 📌 0
A study using an n=1 experiment with single standards at 0V CID reported that 70% of detected ions were in-source fragments (ISFs). www.nature.com/articles/s42... This finding was extrapolated to suggest that ISFs affect all metabolomics experiments to this degree. A counterpoint. rdcu.be/ebFwc 1/n
03.03.2025 20:25 — 👍 30 🔁 15 💬 2 📌 0
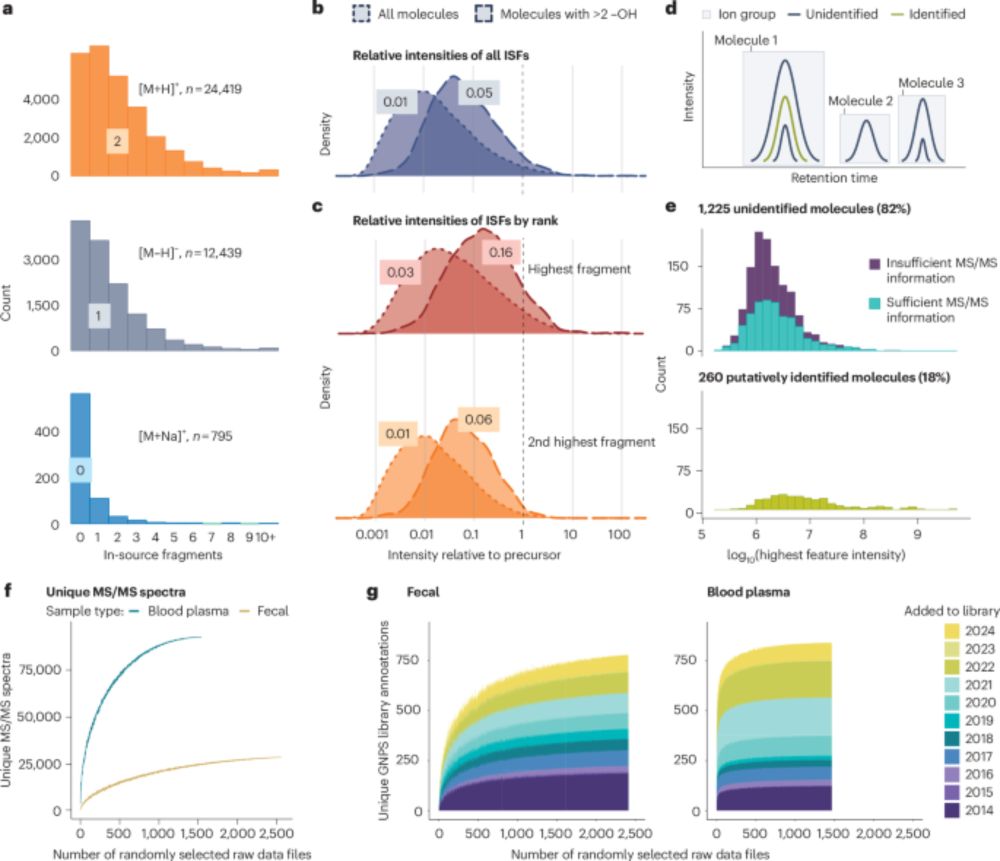
Discovery of metabolites prevails amid in-source fragmentation - Nature Metabolism
Nature Metabolism - Discovery of metabolites prevails amid in-source fragmentation
Excited to share our correspondence in @naturemetabolism.bsky.social, where we address claims that the “dark metabolome” is merely an in‐source fragmentation artifact. www.nature.com/articles/s42... #Metabolomics #TeamMassSpec
03.03.2025 20:24 — 👍 23 🔁 7 💬 1 📌 0
It was a true privilege to have worked on this project! Thanks to @pieterdorrestein.bsky.social, Rob, Rodolfo, @helenamrusso.bsky.social, @simonezuffa.bsky.social, and other co-authors for all the fun we had along the way!
27.02.2025 19:29 — 👍 7 🔁 2 💬 0 📌 0
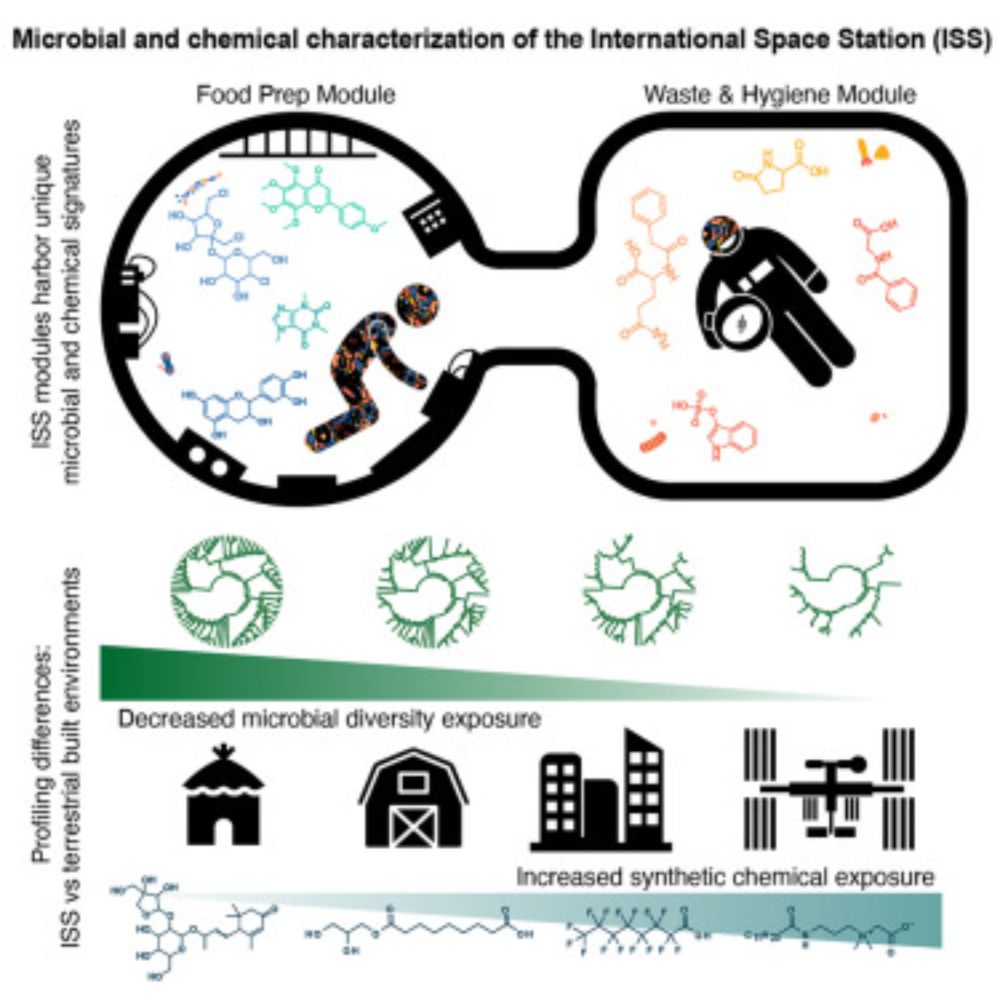
It’s not every day one gets to publish an article with NASA astronauts. This journey started in 2019 and withe the goal to understand the microbial and chemical make-up of the space station. We had lots of experience with analysis from swabs on earth.
27.02.2025 18:52 — 👍 42 🔁 17 💬 1 📌 1

🚨 Abstract deadline alert! 🚨
Friendly reminder to submit your oral abstract before March 6 – don’t miss the chance to showcase your work at hashtag#MetSoc2025!
Registration is now open: lnkd.in/eFZGi4CK
#Metabolomics #EMN
17.02.2025 10:33 — 👍 13 🔁 13 💬 0 📌 0
Our lab at Umeå University uses proteomics to understand protein function and interactions in human gut microbiome species.
Head gut microbiome research @Unilever -opinions are my own-
A 501(c)(3) nonprofit organization that convenes open, peer-reviewed life science meetings over a wide range of fields & global locations.
https://www.keystonesymposia.org/
https://x.com/KeystoneSymp
https://www.youtube.com/KeystoneSymposia
The Huttenhower Lab at the Harvard T.H. Chan School of Public Health: microbial community function and human microbiome population health;
Moderators: @jtnearing.bsky.social, @chahat.bsky.social
Head of Metabolomics at The Francis Crick Institute, London. Made the leap to BlueSky in March 2025 - the air seems cleaner here....
Editor at Nature Microbiology
All opinions are personal
🇩🇪🇸🇪🇮🇳
Strategy & Partnering @Metabolon
Multiomics | Science | Outdoor life
#metabolomics
https://www.linkedin.com/in/timdelany/
microbiome researcher | associate professor @isbscience.org | affiliate prof @uwbioe.bsky.social | ecology, evolution, and systems biology of 💩 🦠 | precision/personalized nutrition/medicine ⚕️ | 🏳️🌈 he/him | lab website: gibbons.isbscience.org
Nature Metabolism publishes studies that advance our understanding of metabolic and homeostatic processes in physiology and disease.
https://www.nature.com/natmetab/
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
Nature Medicine is a research journal devoted to publishing the latest advances in translational and clinical research for scientists and physicians.
www.nature.com/nm
Professor of Microbiome Ecology at Institute of Biology, Leiden University
Bacillus subtilis, biofilms, plant microbiome, bacteria-fungi interactions and circadian clock within @microclockerc.bsky.social #ERCSyG
https://linktr.ee/atkovacs
Scientist Human Gut microbiome @MIB_WUR 🇳🇱 - @DanoneResearch 🇨🇵 - now @RaesLab 🇧🇪 (@VIB_microbes @KU_Leuven). Opinions are my own.
Postdoc UC San Diego | Metabolomics and microbiome
PhD Bioinformatics Wageningen University
Forbes under30 Italy
TEDx speaker
PhD candidate at RECETOX, Faculty of Science, Masaryk University
Working on the profiling of small molecules, particularly #metabolites, in blood based samples using #GC-HRMS, with a focus on #automation of sample preparation
Immunologist and educator; faculty at UVM. Dad; foodie; lover of music.
Professor of Systems Reproductive Medicine,
Director, Robinson Research Institute
and
Co-Director, March of Dimes Prematurity Research Centre at Imperial College London
Trying to understand microbial communities and interactions for a bio-based circular economy at @CMET_UGENT. No tempo libre, @Emigrason_ 🎸
data scientist @ TUM || proteomics, human microbiome, and clinical health research