Just concluded a 2-week coding spree with a student using VS+codex. The x5-10 gain in productivity blew my mind 🤯. BUT, I'm also sure that for inexperienced coders AI-coding makes garbage-in-gargbe-out pitfall inescapable.
31.01.2026 20:21 — 👍 5 🔁 0 💬 0 📌 0
Beautiful “full arc” story on abx response with impressive level of mechanistic detail (could be a class example for undergrad microbiology lesson)
19.01.2026 13:22 — 👍 1 🔁 0 💬 0 📌 0
Congrats on publishing this beautiful work! I'm still mind-blown that: (1) there's a 7th RND pump in E. coli and (2) that its somewhat unclear what it actually pumps out IRL
16.01.2026 18:30 — 👍 2 🔁 0 💬 1 📌 0
It's a beautiful approach! But it's very disappointing that there is actually NO phenotypic characterization of the rearranged strains in the manuscript ... hoping it's an evolving manuscript that they'll keep updating
16.01.2026 18:28 — 👍 0 🔁 0 💬 0 📌 0
Totally agree. Fail early, fail often (and aim high) is something more scientists need to live by.
31.12.2025 02:30 — 👍 1 🔁 0 💬 0 📌 0
Huge credit to @carmenli.bsky.social for her persistence in chasing a moonlight project into its beautiful completion. Credit also goes Ethan Chang, the rotation student contributing to this work 9/9
22.12.2025 15:25 — 👍 1 🔁 0 💬 0 📌 0
Btw, note that “inactivation” can mean more than enzymatic degradation and includes any process that reduces effective drug activity over time (chemical modification, sequestration, etc) 8/9
22.12.2025 15:25 — 👍 1 🔁 0 💬 1 📌 0

We then tested if this assosiation holds through our entire dataset. We used a functional assay for drug inactivation on all drugs and found the association holds up. A long-lag inhibition phenotype is a strong indicator of drug inactivation 7/9
22.12.2025 15:25 — 👍 2 🔁 1 💬 1 📌 0

That pushed us to ask whether cellular defenses might impact curve profiles. We cloned different resistance cassettes and measured how they altered the potency-matched growth curves. This strongly hinted that active drug inactivation underlies a long-lag inhibition profile 6/9
22.12.2025 15:25 — 👍 1 🔁 1 💬 1 📌 0

Overlaying the known mechanisms of action over the barycentric landscape ruled out this effect stem exclusively from how drug target bacteria (since drugs with the same mechanism can land in very different regions of the landscape) 5/9
22.12.2025 15:25 — 👍 1 🔁 0 💬 1 📌 0

Clustering drug by their impact on lag/rate/yield clearly revealed that they vary hugely in how they inhibited growth. In extreme cases, a drug solely affected only a single parameter 4/9
22.12.2025 15:25 — 👍 1 🔁 0 💬 1 📌 0

To compare drugs fairly, we didn’t use an arbitrary concentration. Instead, we interpolated each drug to a potency-matched condition (the concentration expected to produce the same overall level of inhibition) 3/9
22.12.2025 15:25 — 👍 1 🔁 0 💬 1 📌 0

So we assembled a new carefully curated dataset with growth curves across almost forty drugs, measured across multiple sub-inhibitory concentrations. For each curve, we quantified intuitive its key features: lag, growth rate, and yield 2/9
22.12.2025 15:25 — 👍 2 🔁 1 💬 1 📌 0

Predicting drug inactivation by changes in bacterial growth dynamics - npj Antimicrobials and Resistance
npj Antimicrobials and Resistance - Predicting drug inactivation by changes in bacterial growth dynamics
Our recent paper in npj Antimicrobials and Resistance is a great example of scientific serendipity: after staring at thousands of bacterial growth curves over many studies, we started wondering whether the curve shapes themselves carry mechanistic information 1/9 🦠🧪
www.nature.com/articles/s44...
22.12.2025 15:25 — 👍 44 🔁 25 💬 2 📌 2
This was a true collaboration between physicists (Andrew Mugler and @motasemelgamel.bsky.social), an immunologist (Michael Brehm from @umasschan.bsky.social ), and systems biologists (with @serkansayin.bsky.social and Brittany Rosener from my lab also at @umasschan.bsky.social ) (7/7)
17.12.2025 18:52 — 👍 2 🔁 0 💬 0 📌 0

Beyond providing (to our knowledge) the first dynamical model for tumor colonization, our study matters given the fierce debate on the tumor microbiome. These statistical “fingerprints” may help distinguish genuine colonizers from technical artifacts/contamination (possibly even by microscopy) (6/7)
17.12.2025 18:52 — 👍 2 🔁 1 💬 1 📌 0

The surprise: lineage sizes formed a scale-free power law that matches Zipf’s law (rank–frequency slope ~−1). This signature was robust across dozens of tumors and multiple collection days post bacteria intratumor injection (5/7)
17.12.2025 18:52 — 👍 2 🔁 0 💬 1 📌 0

When we injected bacteria directly into the tumor (circumventing the bottleneck), we detected thousands of colonizing lineages, yet their sizes were still highly uneven (ruling out early tumor arrivers dominate) (4/7)
17.12.2025 18:52 — 👍 1 🔁 0 💬 1 📌 0

Since we used genetically barcoded bacteria, we could also monitor growth of individual colonizers. We found that growth was extremely uneven with a handful of lineages becoming dominant (“winner-takes-most”) (3/7)
17.12.2025 18:52 — 👍 1 🔁 0 💬 1 📌 0

Main takeaways: Post systemic infection, there's a tight colonization bottleneck (per-cell colonization probability ~0.005%). Yet, once colonization happens, growth is remarkably fast (~50 min generation time) and bacterial load in tumors approaches saturation within a day (2/7)
17.12.2025 18:52 — 👍 1 🔁 0 💬 1 📌 0

We just published in @molsystbiol.org with the Mugler lab (UPitt) on bacterial population dynamics during tumor colonization (mouse model). Our study was guided by a Luria–Delbrück-style idea: infer mechanism from statistics (1/7) 🧪🦠
doi.org/10.1038/s443...
17.12.2025 18:52 — 👍 16 🔁 11 💬 2 📌 0
Great opportunity to participate in scientific outreach!
22.05.2025 18:05 — 👍 0 🔁 0 💬 1 📌 0
YouTube video by MicrobeTV
Matters Microbial #89: Can AI Point Us to New Antibiotics
Had a fantastic time with @markowenmartin.bsky.social’s on his #MattersMicrobial podcast (@microbetv.bsky.social). We discussed how to uncover the killing mechanisms of hundreds of antibacterials simultaneously and how #AI can transform the search for new antibiotics 🧪🦠💊
youtu.be/Wa0zd-TRCO8?...
05.05.2025 16:47 — 👍 5 🔁 2 💬 1 📌 0
Yo, micro/evo/qbio/physics of living systems/astrobiology/renegade cell bio folks- if you are interested in starting a feed, reply/repost this. Let's get the crew back together!
29.01.2025 23:40 — 👍 26 🔁 7 💬 5 📌 0
Fascinating shift in the geography of drug development (excellent writing too!)
26.01.2025 03:29 — 👍 0 🔁 0 💬 0 📌 0
The inherent stochasticity of biological systems 😉
18.01.2025 19:46 — 👍 1 🔁 0 💬 0 📌 0
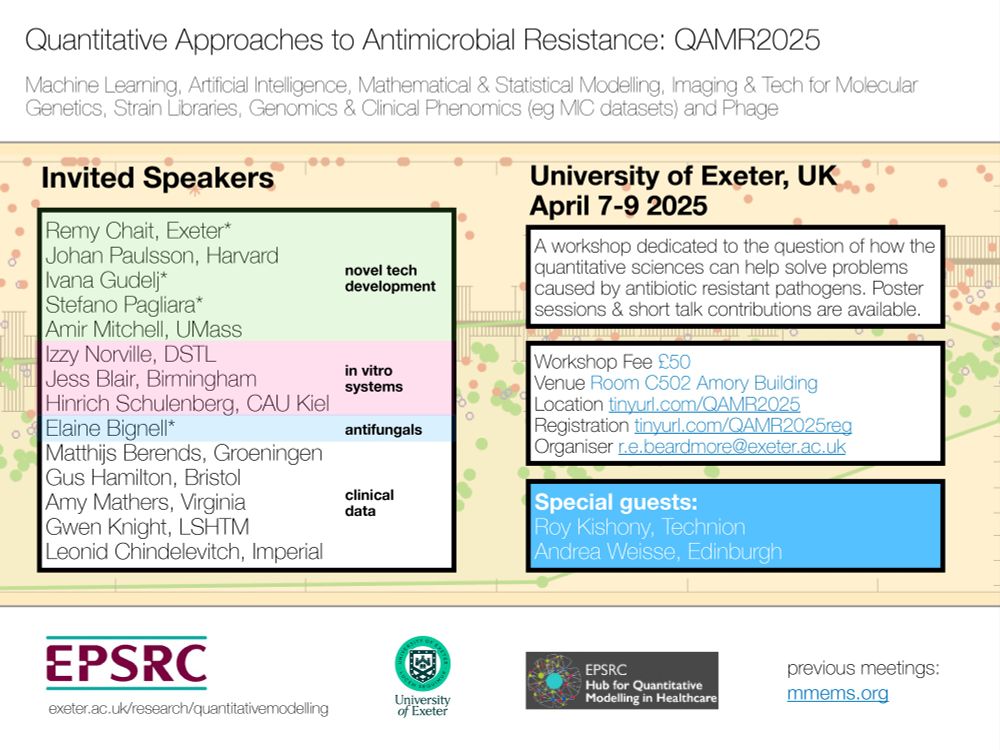
Interested in Antimicrobial Resistance & Quantitative Bio? Join a 3-day meeting+workshop with some of my favorite #AMR #QBio scientists this April (University of Exeter, UK). Register here: tinyurl.com/QAMR2025reg 🧪🦠
17.01.2025 15:16 — 👍 5 🔁 1 💬 0 📌 0
Amazing book, shaped my worldview on Science. Kuhn’s genius was also in claiming that “paradigm shifts” are not purely logical, but are tainted by social & psychological factors. He claimed that established scientists are often conservatives resisting such shifts.
28.12.2024 03:02 — 👍 6 🔁 0 💬 0 📌 0
It was all code written in Matlab
14.12.2024 19:50 — 👍 0 🔁 0 💬 0 📌 0
Scientist fascinated by infectious diseases, host-microbe interactions and bacterial infections. Views my own 🇵🇷 #CienciaBoricua #MacFellow #MRSA
Welsh postdoc @ The Crick with Dr. Eachan Johnson.
TB and ESKAPE pathogen research. Interested in high-throughput, genomics, and chem biol.
PhD from University of Birmingham with Prof. Jess Blair.
Views my own. (she/her)
Professor of Antimicrobial Resistance. Institute of Microbiology and Infection, University of Birmingham. Director University of Birmingham Doctoral School. Editor in Chief of npj Antimicrobials and Resistance.
Just another LLM. Tweets do not necessarily reflect the views of people in my lab or even my own views last week. http://rajlab.seas.upenn.edu https://rajlaboratory.blogspot.com
We are involved in understanding the ecological and evolutionary forces that shape microbial communities in nature.
Visit our website - lifewp.bgu.ac.il/wp/imizrahi/
Twitter - https://x.com/LabMizrahi
Bacteria lover. Good, bad, or other wise. Beneficial, commensal, or pathogenic alike.
Authorized GMB (Genetically Modified Bacteria) scientist
Precision-function-based microbiome trials.
Ex vivo microbiome assays.
Microbiome–xenobiotic interactions.
Meta-omics analyses
Professor and Chair, Department of Microbiology, University of Chicago. Study the pathogenesis of tuberculosis, and other mycobacterial infections. Opinions my own.
Single cell systems and synthetic biology lab at Northwestern University and Chan Zuckerberg Biohub Chicago.
https://www.goyallab.org/
Get more out of large conferences with our Mentoring and Networking program.
https://binningsingletons.com/
https://journals.asm.org/doi/10.1128/msphere.00643-19
Senior Research Associate at MRC Toxicology, University of Cambridge
My research interests focus on interspecies-interactions in gut and food microbial communities, their effects and benefits, as well as bacterial responses to drugs and food compounds.
Computational microbiologist
Senior Lecturer, Newcastle Uni UK
We study the chemical language of bacteria (& what influences it) for antibiotic discovery
http://www.medicinesfromthesea.com
Editor Microbiology, Young Academy of Scotland, EAB RSC NatProdRep & EAB ACS JNatProd
Professor of Microbiome Ecology at Institute of Biology, Leiden University
Bacillus subtilis, biofilms, plant microbiome, bacteria-fungi interactions and circadian clock within @microclockerc.bsky.social #ERCSyG
https://linktr.ee/atkovacs
#SynBio and #naturalproducts. Enzymology, heterologous expression, and bioengineering. Senior Lecturer in Chemical and Synthetic Biology
at the University of Sydney
.
Swimming🏊♀️ sewciophilic🧵microbial 🧫biochemist🧪🧬/biomaterials ⚒️ scientist;
(bio)chemistry, materials and ecobiology of actinos/natural products;
microbiology/chemistry in hot tubs;
Biofilm Alliance;
Advisory Council member Microbiology Society;
She/her
Natural products biosynthesis and computational biologist at JGI. Host of http://naturalprodcast.com This is my personal account, not my employer's, and things I say or re-post are not representative of them, and maybe not even me.
Microbiology and bioinformatics professor in Tuebingen, Malinois lover, fan of secondary metabolites in bacteria, evolution, computational biology, yoga, search and rescue, and Pepper
Microbiome gen(om|et)ics, secondary metabolism, comp bio, antibiotic/drug disco, evolution, & hockey. he/him.