Microbial Databases, Software Tools, and Web Services
Publish your microbial database or software tool in MRA to gain recognition, citations, and community trust.
Excited to share our new call for papers at @asm.org's MRA journal. Do you have a database, software, or web-based tool that would be a great resource for the micro community? Consider submitting it to MRA! journals.asm.org/journal/mra/...
23.07.2025 15:38 — 👍 29 🔁 20 💬 0 📌 0
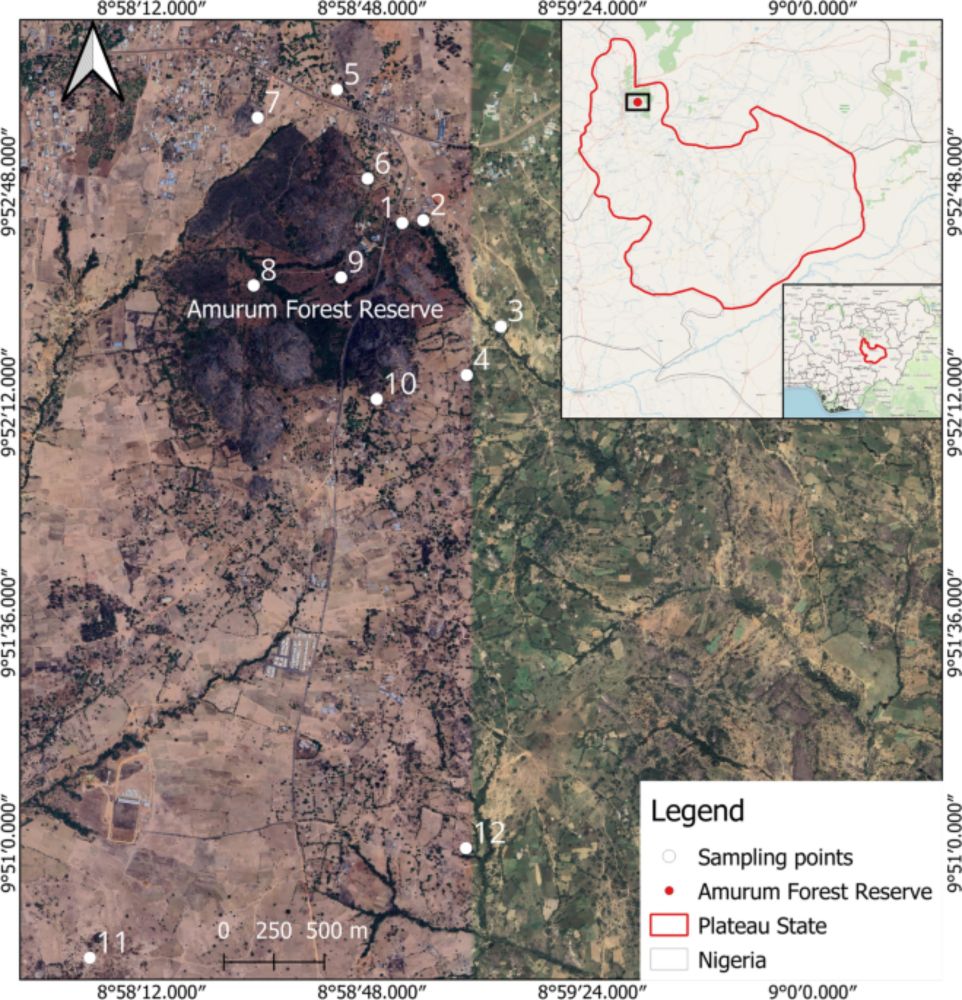
First detection of Sindbis virus in wild birds in Nigeria - Scientific Reports
Scientific Reports - First detection of Sindbis virus in wild birds in Nigeria
Our latest paper in Scientific Reports: First detection of #Sindbis #virus in wild birds in Nigeria! Screened 504 birds, found SINV RNA in African Thrushes, sequenced genome linking to Algeria, Spain & Kenya. Calls for enhanced surveillance & One Health 🦜🦟 doi.org/10.1038/s415...
10.07.2025 13:40 — 👍 1 🔁 0 💬 0 📌 0

Back a London Calling 10 years after my first #NanoporeConf
21.05.2025 15:34 — 👍 4 🔁 2 💬 0 📌 0
Not a single marine species among the 50! most studied bacteria. Gobsmacking.
Ten species comprise half of the bacteriology literature, leaving most species unstudied www.biorxiv.org/content/10.1...
06.01.2025 09:30 — 👍 36 🔁 7 💬 3 📌 3
Does anyone have updated benchmarks for #Dorado basecalling on different GPUs, including Apple Silicon? #Nanopore #Sequencing
26.12.2024 10:38 — 👍 9 🔁 1 💬 0 📌 0

Minion Mk1D
Happy to receive a Christmas gift before the holidays #nanopore #minion #Mk1D
12.12.2024 13:54 — 👍 7 🔁 1 💬 0 📌 0
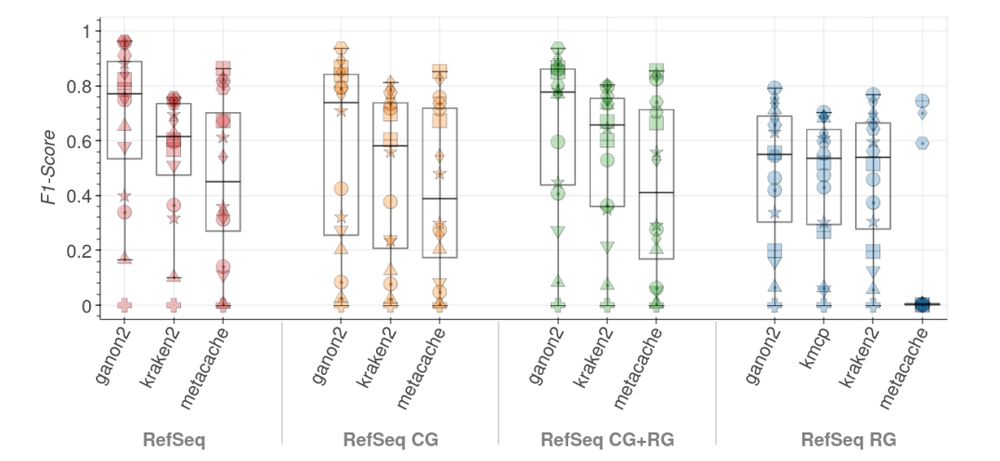
Source:
https://www.biorxiv.org/content/10.1101/2023.12.07.570547v2.full.pdf
ganon2: up-to-date and scalable metagenomics analysis. #Metagenomics #Genomics #Bioinformatics 🧬 🖥️ @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
11.12.2024 19:05 — 👍 7 🔁 1 💬 0 📌 0
15th CBRNe Protection Symposium and the Exhibition of CBRNe protection equipment – Welcome to The 15th CBRNe September 30 – October 2, 2025 in Malmö, Sweden
Sign up for next year's CBRNe Protection Symposium! A perfect opportunity to learn more about the latest in CBRNe protection research, showcase your own research. We also welcome abstract submissions for oral or poster presentation. More information here: ow.ly/g2kv50Ui2Hx
Via @FOIresearch on X
06.12.2024 10:09 — 👍 1 🔁 0 💬 0 📌 0
Working on designing a new hybridisation capture panel for a virus family. Previously used CATCH to select optimal set of probes but now looking for alt tools setting tiling coverage to 2 or 3 intead of complete coverage. Any recommendation of tools or code that can do it? #bioinformatics
06.12.2024 09:56 — 👍 0 🔁 0 💬 0 📌 0

Promotional logo for MMseqs2 16 with the MMseqs2 Rocket mascot as a smart phone like App logo
MMseqs2 Release 16 Highlights: GPU-accelerated search📄, ORF or new 6-frame translated search modes, contig taxonomy always keeps the longest ORF, bug fixes (reduced memory and higher sensitivity) and relicensed as MIT
📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶
27.11.2024 09:08 — 👍 116 🔁 44 💬 0 📌 1
Please share! Applications due Dec 15th for a starting date in Sep 2025.
26.11.2024 17:48 — 👍 15 🔁 20 💬 0 📌 0

Microbiology Resource Announcements Open Call for Reviewing Editors
Microbiology Resource Announcements is looking for microbiologists with expertise in genome sequencing to join our Board of Reviewing Editors.
We're looking for Reviewing Editors, over at Microbiology Resource Announcements (MRA) - call in the link below.
It's an ideal position for early-career researchers looking for a steady editing role.
#microbiology #editing #publishing #scipub #genomics #sequencing
journals.asm.org/mra/open-cal...
15.11.2024 08:53 — 👍 1 🔁 2 💬 0 📌 0
Spending my afternoon installing Ubuntu on my old MacBook Pro 2012. Hopefully it will be a good complement to my M3 MacBook
04.11.2024 15:37 — 👍 0 🔁 0 💬 0 📌 0
I'm going to be advertising a PhD studentship (open to international students) working on long read pangenome / assembly algorithms in bacteria. Would suit someone with maths/compsci background. Aptitude and interest more important than experience. Do please spread the word. Advert to follow
22.10.2024 17:56 — 👍 33 🔁 50 💬 1 📌 2
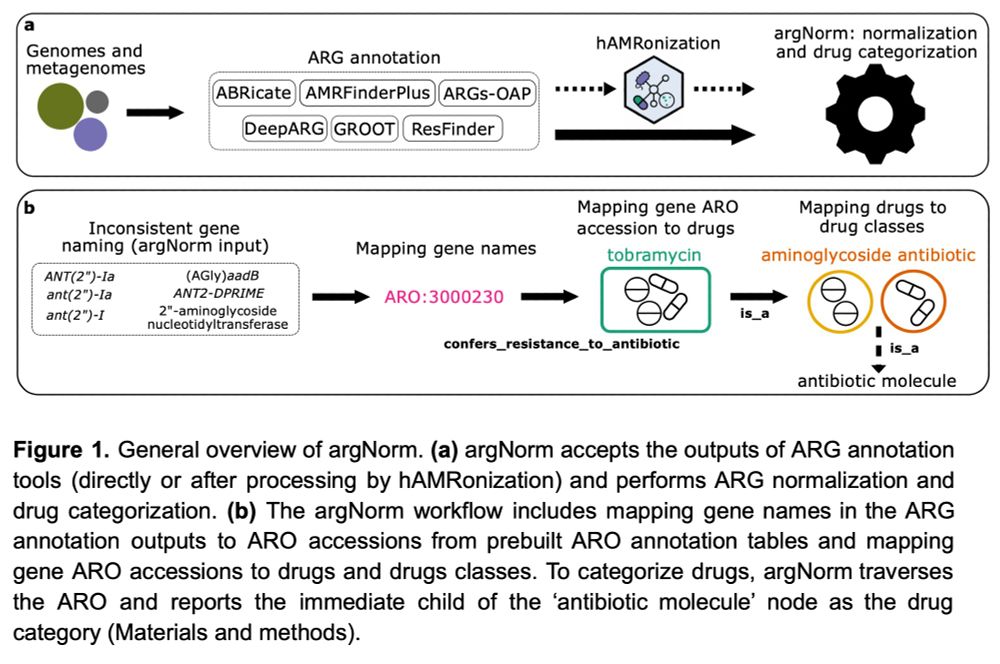
argnorm
Hey #AMR people,
the argNorm preprint is now available on #QUT ePrints: eprints.qut.edu.au/252448
We* designed a tool for normalizing ARG annotations across currently popular tools & dbs.
*@svetlanaup.bsky.social
Vedanth Ramji
Hui Chong
Yiqian Duan
Finlay Maguire
@luispedrocoelho.bsky.social
1/6
10.10.2024 07:56 — 👍 14 🔁 11 💬 1 📌 3
Happy to assist if you need help.
22.10.2024 17:30 — 👍 1 🔁 0 💬 1 📌 0
Did they solve your Ftul problem?
22.10.2024 16:53 — 👍 1 🔁 0 💬 1 📌 0

Genomics+Bioinformatics Starter Pack 🧬🖥️
Join the conversation
I created a genomics+bioinformatics starter pack. If I left you off, *please* reply and I'll add you! go.bsky.app/B5YYBfq
22.10.2024 12:34 — 👍 176 🔁 106 💬 99 📌 7
Last chance to fill in our survey! Have your say while you can :) Thanks for all those of you that already have!
21.10.2024 03:46 — 👍 13 🔁 11 💬 1 📌 0

Lets start the scientific evaluation of different Semla buns
13.02.2024 16:56 — 👍 2 🔁 0 💬 0 📌 0
A metagenomic alpha-diversity index for microbial functional biodiversity academic.oup.com/femsec/artic... #jcampubs
12.02.2024 14:12 — 👍 3 🔁 2 💬 0 📌 0

PLA-complexity of k-mer multisets
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
Md. Hasin Abrar and I recently posted a pre-print "PLA-complexity of k-mer multisets" We explore a measure of a k-mer multiset complexity, first suggested by Boffa et al, which we call the PLA-complexity.
doi.org/10.1101/2024...
(1/n)
13.02.2024 07:47 — 👍 5 🔁 4 💬 1 📌 0
Nearly 20 years ago, Jill Banfield, Gene Tyson & team transformed #microbiology with their seminal paper on genome-resolved metagenomics. Their innovative approach in reconstructing microbial genomes opened new frontiers in understanding ecosystem dynamics!
paper here: www.nature.com/articles/nat...
31.01.2024 16:59 — 👍 14 🔁 6 💬 0 📌 1
I make stuff to help scientists focus on their research david4096.github.io
Computational biology and bioinformatics, artificial intelligence, tango and literature are some of my interests, CNRS - Bordeaux. Opinions are my own
Professor at Cornell Univ. Bacillus subtilis physiology and stress responses (metal homeostasis, redox stress, antibiotic resistance & cell envelope). EIC of Mol Microbiol.
A biotech engineer with a fascination for evolution and molecular bio.
Computational Biologist turned AI and machine learning enthusiast. Interested in any topic ending in -omics, with a focus on predicting cancer outcomes, disease mechanisms, and protein function.
Computational Biologist
#PhD #plant_biotech
https://orcid.org/0000-0002-0815-9986
Undergraduate in bioinformatics 🫶 | Microbiology Lab research assistant | Here to grow, learn, and contribute ✨
Metagenomics bioinformatics analyst Minnesota supercomputing institute also like bears, 3d printing, tabletop gaming.
Postdoc in computational biology/bioinformatics
Specialist Director
Norwegian Institute of Public Health.
Personal posts, not employer’s views.
https://orcid.org/0000-0001-5909-7917
AMR Antimicrobial Resistance
Fernanda C. Petersen,
Professor, University of Oslo and
Ulf R. Dahle,
Specialist Director,
Norwegian Institute of Public Health
Human genetics; epigenetics; infectious disease; ageing; dementia. The usual stuff. Oh, Public Health too. Faculty at the University of Pittsburgh.
Professor in animal breeding at Swedish University of Agricultural Science. Also Pro Vice-Chancellor with reposibility for research infrastructure. Will write about research and academia. opinions are my own!🧬🧪🐄🐷🐔🥚🐟
Our electrophoresis and PCR systems are designed for teaching hands-on labs in classrooms. MiniOne Systems ensures a seamless transition to NGSS.
https://orcid.org/0000-0003-2815-1735
Long read (DNA, RNA) | Epigenetics (bacteria, human) | Professor of Genomics @IcahnMountSinai | Humid Lab🧫💻🧪 http://fanglab.bio