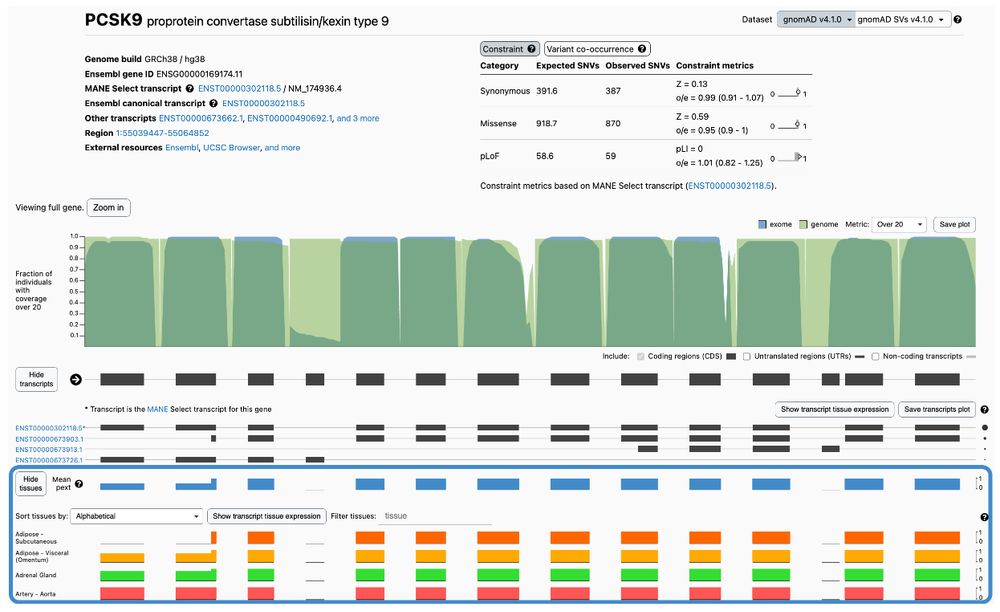
Proportion expressed across transcripts (pext), using GTEx v10, is now available on #gnomAD v4!
22.11.2024 14:38 — 👍 15 🔁 3 💬 0 📌 3@gnomad-project.bsky.social
The world's largest open resource of human genetic variation. For help please use http://broad.io/gnomad_forum; feature requests/bug reports to http://broad.io/gnomad_github

Proportion expressed across transcripts (pext), using GTEx v10, is now available on #gnomAD v4!
22.11.2024 14:38 — 👍 15 🔁 3 💬 0 📌 3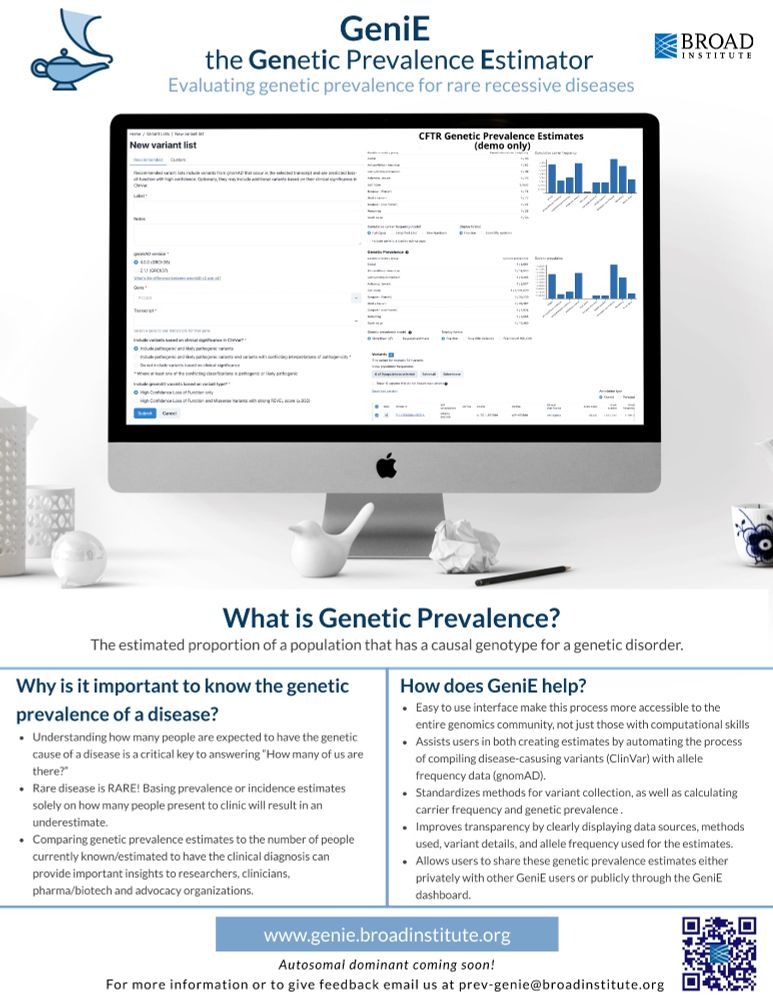
GeniE, the genetic prevalence estimator, is now available! broad.io/genie
This tool allows users to estimate the genetic prevalence of autosomal recessive diseases using #gnomAD allele frequency data & classifications from #ClinVar
Blog post: broad.io/genie_blog
gnomAD 4.1 is now live! This release fixes the AN issue in #gnomAD v4.0 & adds 2 new functionalities:
1) Joint AN across all called sites in exomes and genomes
2) A flag indicating when exomes and genomes frequencies are highly discordant
Learn more at broad.io/gnomad_v4-1
A special congratulations to Konrad Karczewski, Siwei Chen, @ksamocha.bsky.social and Mike Guo for all their hard work on these publications. Read more about this work at www.nature.com/articles/d41... (2/2)
06.12.2023 18:21 — 👍 3 🔁 1 💬 0 📌 0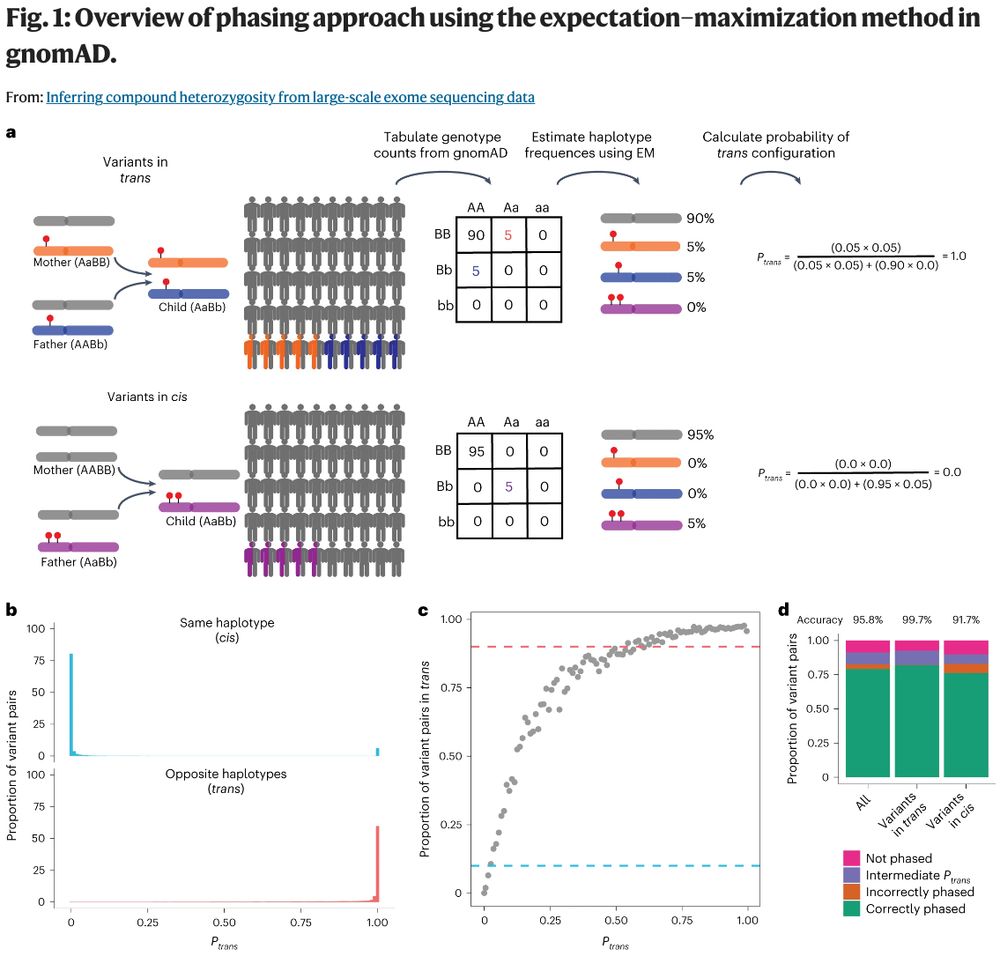
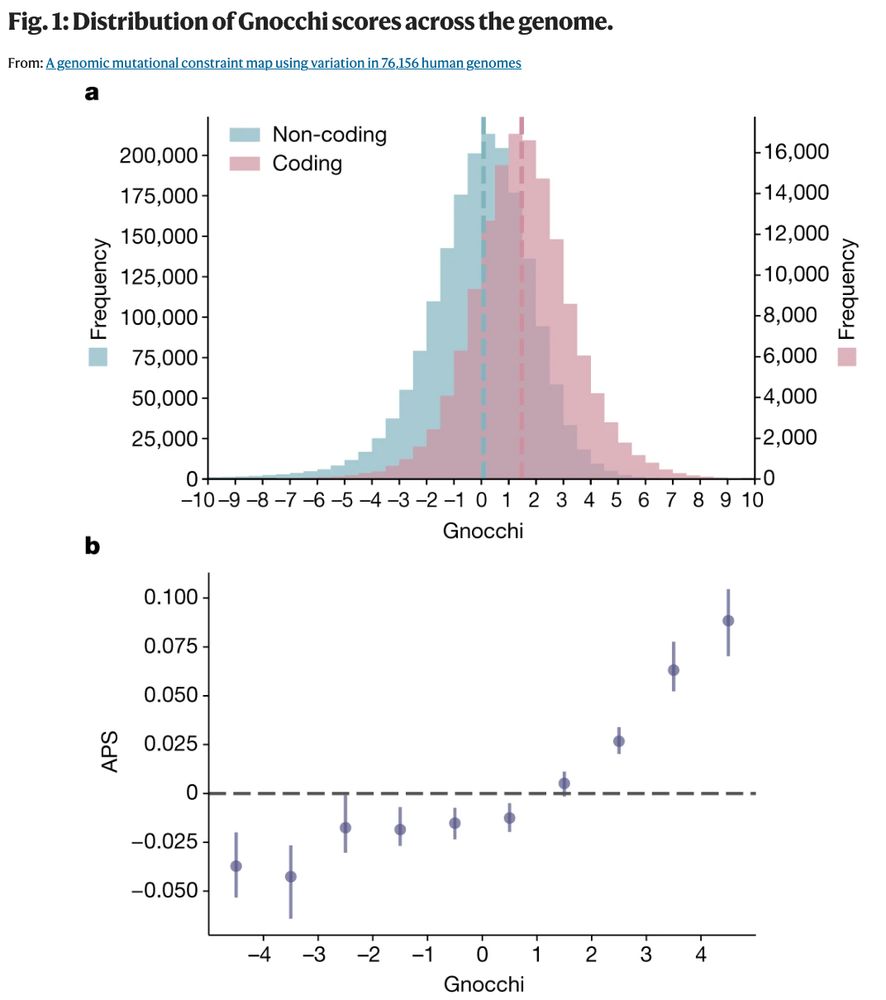
The #gnomAD v3 papers are now published! This includes the non-coding constraint paper broad.io/gnomAD_v3_non_coding & inferring compound heterozygosity paper broad.io/gnomAD_v3_comp_het Congratulations to everyone who contributed to this valuable work! (1/2)
06.12.2023 18:19 — 👍 2 🔁 0 💬 1 📌 0As part of v4, we are happy to announce the launch of the #gnomAD forum broad.io/gnomad_forum. This will be a place for our users to help each other, discuss the data and ask questions. #ASHG2023
05.11.2023 14:01 — 👍 1 🔁 0 💬 0 📌 0To learn more about what is involved with QCing the gnomAD v4 dataset please attend Julia Goodrich’s #ASHG2023 talk today (11/4) at 10:30am in ballroom B.
04.11.2023 13:00 — 👍 1 🔁 1 💬 1 📌 0Gene constraint is now available on #gnomAD v4! This is the first time we have had constraint data available on GRCh38. Katherine Chao will be covering this work during her talk at #ASHG23 tomorrow (11/4) at 11am in rm 202A.
03.11.2023 21:36 — 👍 6 🔁 3 💬 0 📌 1To learn more about the impact of diversity on variant discovery and gene constraint please attend Katherine Chao’s #ASHG23 talk tomorrow (11/4) at 11am in rm 202A
03.11.2023 13:01 — 👍 4 🔁 2 💬 0 📌 0Keep an eye on this account for the next few days to learn more about our methods including structural variation, genomic ancestry, and our QC process (11/11)
02.11.2023 13:51 — 👍 0 🔁 0 💬 0 📌 0And of course, a special thank you to the 308 gnomAD data contributors and all the individuals who have enrolled in research. Without their willingness to share data and participate in research, gnomAD would not exist! gnomad.broadinstitute.org/about (10/11)
02.11.2023 13:51 — 👍 0 🔁 0 💬 1 📌 0
The full gnomAD v4 launch team was composed of multiple groups at the Broad spanning the MPG and DSP programs as listed below, with most data generated by @BroadGenomics. Thank you!!! (9/11)
02.11.2023 13:50 — 👍 0 🔁 0 💬 1 📌 0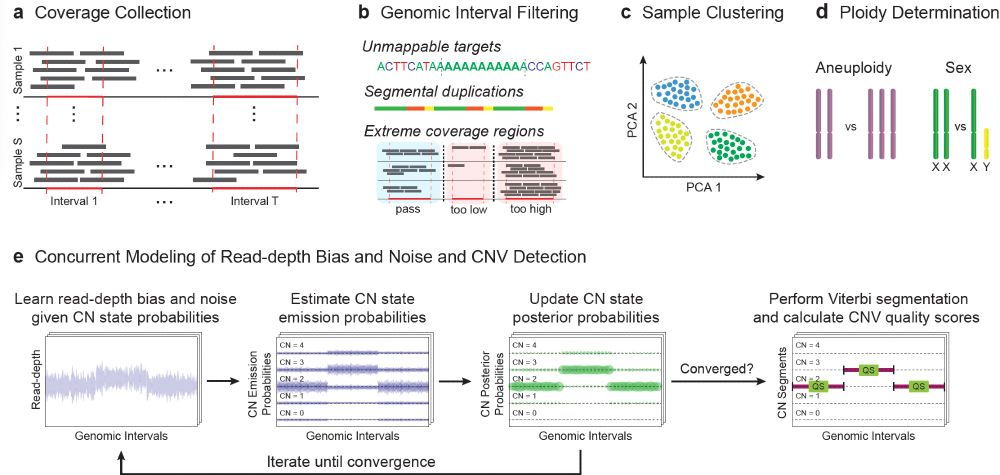
To learn more about the technical details of how this data was generated read our blog posts broad.io/gnomad_v4_sv and broad.io/gnomad_v4_cnv. If you are attending #ASHG23 please attend Jack Fu’s talk today (11/2) at 1:45pm in rm 202A (2/2)
02.11.2023 12:58 — 👍 1 🔁 0 💬 0 📌 0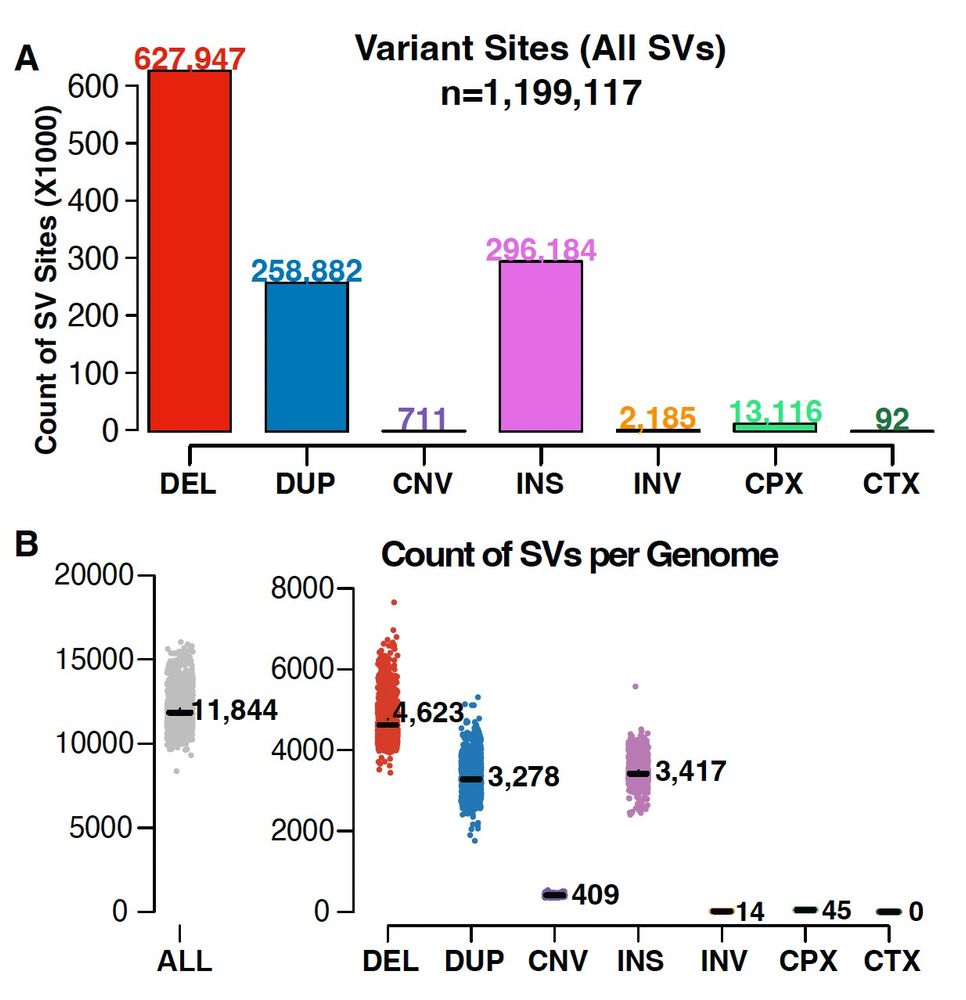
As part of #gnomAD v4, in collaboration with the Talkowski Lab, we have released 1,199,117 genome SVs and 66,903 rare exome CNVs. These data represent the first gnomAD SV dataset released native to the GRCh38 reference genome. (1/2)
02.11.2023 12:58 — 👍 7 🔁 3 💬 1 📌 0A special thanks to the hail team (hail.is), especially Tim Poterba, Chris Vittal, Dan King, Jackie Goldstein, and Daniel Goldstein for all their work on this release. (8/11)
01.11.2023 18:38 — 👍 0 🔁 0 💬 1 📌 0Thank you to our browser team, especially @msol.bsky.social, Phil Darnowsky, Riley Grant, @stephenjahl.bsky.social and Elissa Alarmani, who put in long hours and late nights these last few weeks to get us over the line! (7/11)
01.11.2023 18:37 — 👍 0 🔁 0 💬 1 📌 0We would like to thank the entire gnomAD Production Team broad.io/gnomad_team especially Julia Goodrich, Katherine Chao, Mike Wilson, Kristen Laricchia, and @sambaxter.bsky.social who did a massive push over the last few months to release v4 in time for #ASHG23 (6/11)
01.11.2023 18:36 — 👍 1 🔁 0 💬 1 📌 0If you are attending #ASHG23 please stop by the @BroadInstitute Clinical Lab booth (#938) during our #gnomAD office hours to meet the team, ask questions, and learn more about v4. (5/11)
11/2 & 11/3: 10-10:45am, 3-5pm
11/4: 10-10:45am, 2:15-4:15pm
#ASHG2023
Our genetic ancestry blog broad.io/gnomad_ancestry discusses our efforts to improve representation in #gnomAD, how we label groups and how the diversity in gnomAD is improving genomic filtration. (4/11)
01.11.2023 18:35 — 👍 0 🔁 0 💬 1 📌 1QC of v4 required analyzing over 1 BILLION variants! More than 910 million variants passed our filters and are available on our browser broad.io/gnomad. More details on this variant dataset are available on our new stats page broad.io/gnomad_stats (3/11)
01.11.2023 18:35 — 👍 0 🔁 0 💬 1 📌 1Development of v4 involved jointly calling almost 1 million samples, which is the largest callset ever made at the @broadinstitute. Working at this scale required extensive collaboration across the Broad. Learn what went into making v4 read our blog broad.io/gnomad_v4 (2/11)
01.11.2023 18:35 — 👍 0 🔁 0 💬 1 📌 0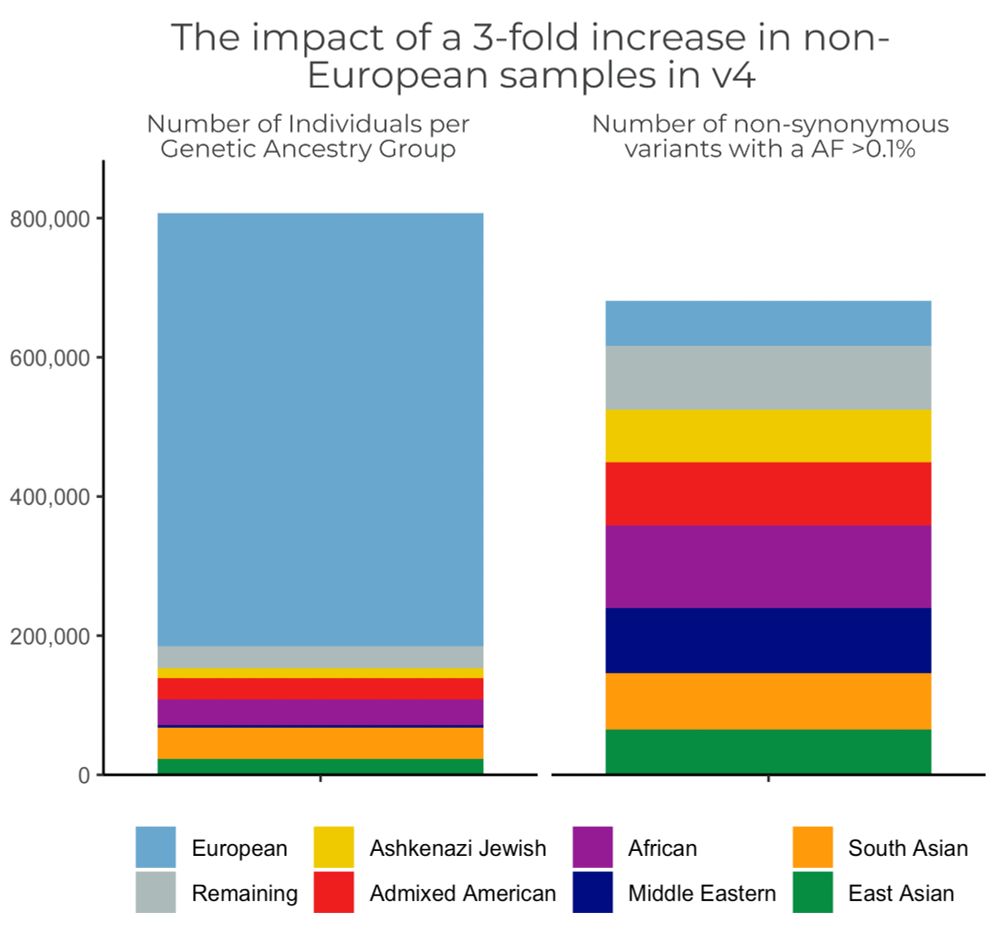
The #gnomAD team is proud to announce the release of gnomAD v4! The v4 dataset includes 730,947 exomes & 76,215 genomes, which is ~5x larger than the v2 & v3 releases combined, & includes nearly 120K indivs of non-European genetic ancestry broad.io/gnomad #ASHG23 (1/11)
01.11.2023 18:35 — 👍 32 🔁 21 💬 2 📌 2