
Scientist on Friday, superfan on Sunday. Balance. 😌
16.11.2025 16:30 — 👍 5 🔁 1 💬 0 📌 0@aakashg20.bsky.social
Grad Student - University of Pittsburgh Arndt Lab - www.karenarndtlab.com UG - Shiv Nadar University Epigenetics and Gene Expression (He/Him/His) 🏳️🌈

Scientist on Friday, superfan on Sunday. Balance. 😌
16.11.2025 16:30 — 👍 5 🔁 1 💬 0 📌 0Delhi Crime Season 3🤯
16.11.2025 22:16 — 👍 0 🔁 0 💬 0 📌 0
Beautiful review from J. Rudolph on Histone PARylation factor 1: a review of its role in the DNA damage response url: academic.oup.com/nar/article/...
11.11.2025 14:49 — 👍 20 🔁 6 💬 1 📌 1Dead Poets Society🤌🥲
09.11.2025 06:12 — 👍 0 🔁 0 💬 0 📌 0I love AlphaFold—but please include PAE (Predicted Aligned Error) plots for every “interaction.” Pretty PDBs ≠ proof. If the PAE doesn’t show an interface, it ain’t one. Show the plots. Structural biology's reputation is on the line.
24.10.2025 17:44 — 👍 127 🔁 29 💬 5 📌 4Just submitted this a few hours ago. We ❤️ biorxiv. This is work from my student Payal Arora who will defend her thesis very soon!
24.10.2025 21:24 — 👍 35 🔁 8 💬 0 📌 0I don’t think this is a controversial opinion but …
Indian style mutter-paneer definitely deserves the Nobel peas prize this year
www.indianhealthyrecipes.com/matar-paneer...
We have added additional functionalities including a collapsible protein pane and zoom scrolling. domaindesigner.farnunglab.com
04.10.2025 19:19 — 👍 18 🔁 6 💬 1 📌 1
Protein Domain Designer tool for the generation of publication-ready protein domain diagrams
New week, new tool: Find our Protein Domain Designer tool to generate publication-ready protein domain diagrams here: domaindesigner.farnunglab.com
29.09.2025 15:01 — 👍 186 🔁 66 💬 1 📌 3
The histone chaperone Spt6 controls chromatin structure through its conserved N-terminal domain
22.09.2025 11:33 — 👍 5 🔁 1 💬 0 📌 0😂😂😂
27.09.2025 22:05 — 👍 0 🔁 0 💬 0 📌 0
This is a fascinating paper that particular types of RNA binding proteins with IDRs that target nuclear speckles also can recruit their own RNAs to nuclear speckles as a negative feedback mechanism for condensation the authors call "interstasis" www.nature.com/articles/s41...
25.09.2025 20:24 — 👍 30 🔁 11 💬 0 📌 0
This Wednesday at #FragileNucleosome seminar, we are excited to host Ali Wilkening and @schvartzman.bsky.social to tell us about amazing work they are doing!
Register here for upcoming session and the entire series:
us06web.zoom.us/webinar/regi...

New preprint is up, led by former postdoc Braulio Bonilla.
We know these remodelers regulate nucleosome positions and occupancy, but do they have a role in regulating higher order chromatin structure? We take a peek for BAF and INO80C.
www.biorxiv.org/content/10.1...
Fantastic preprint from my colleague at Pitt. Was presented at recent CSH meeting #moet2025 #moet
03.09.2025 21:23 — 👍 7 🔁 1 💬 0 📌 0
Had the most amazing time at Cold Spring Harbor Laboratory! #moet2025
02.09.2025 00:24 — 👍 2 🔁 0 💬 0 📌 0
Fragile Nucleosome Fall Seminars: September 10 Grace Bower, Kvon Lab @ UC Irvine, USA Jennifer Phillips-Cremins, UPenn, USA September 24 Ali Wilkening, Sanulli Lab @ Stanford, USA Juanma Schvartzman, Columbia University, USA October 8 Sanim Rahman, Greenberg Lab @ UPenn, USA Alex Federation, Talus Bio, USA October 22 Alice Laigle, Croll Lab @ University of Neuchâtel, Switzerland Seungsoo Kim, UC Irvine, USA November 5 Hannah Long, University of Edinburgh, UK Jeff Vierstra, Altius Institute, USA November 19 Ishtiaque Hossain, Pastor Lab @ McGill, Canada Sarah Teichmann, University of Cambridge, UK December 3 María Mariner Faulí, Rada Iglesias Lab @ IBBTEC, Spain Jonathan Henninger, Carnegie Mellon, USA December 17 Rebecca Berrens, Oxford University, UK Jean-Benoit Lalanne, University of Montreal, Canada
We're super excited to announce the entire lineup for the Fall season of Fragile Nucleosome Seminars, starting on Sept 10th at 1200 EDT / 1600 UTC with @gracebower.bsky.social and @creminslab.bsky.social!
register here for the entire series: us06web.zoom.us/webinar/regi...

Our collaborative paper with the Arndt lab is out today: doi.org/10.1093/nar/... (originally bioRxiv: www.biorxiv.org/content/10.1...). Here we asked the question of how CHD1 localizes to chromatin.
28.08.2025 23:07 — 👍 36 🔁 10 💬 1 📌 0🧬 Transcription elongation by RNA polymerase II relies on a web of elongation factors. Our new work shows how IWS1 acts as a modular scaffold to stabilize & stimulate elongation. Fantastic work by Della Syau! www.biorxiv.org/content/10.1...
29.08.2025 10:14 — 👍 56 🔁 23 💬 1 📌 1
Final version of Bingbing Duan’s main thesis work is out now now in Nature Communications @natureportfolio.nature.com www.nature.com/articles/s41... I’m so pleased with this study that we’ve been working on for so long #epistasis #transcription #geneexpression 1/
28.08.2025 01:33 — 👍 28 🔁 15 💬 1 📌 0
To all post-docs: The Genome Biology dept @embl.org
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

Two shy dinos caught on the kiss cam! 📸
26.07.2025 15:26 — 👍 6 🔁 1 💬 0 📌 0I am pleased to announce Reactor—the complex formation calculator for multi-component protein complex formation experiments. Developed by the Farnung Lab @harvardmed.bsky.social.
reactor-farnunglab.pythonanywhere.com 1/n
New paper:
More than 2700 human 3′UTRs are highly conserved. These 3′UTRs are essential components in mRNA templates, as their deletion decreases protein activity without changing protein abundance. Highly conserved 3′UTRs help the folding of proteins with long IDRs.
www.biorxiv.org/content/10.1...
Yes, yes, and yes!
www.nature.com/articles/d41...

www.biorxiv.org/content/10.1... - Awesome!
03.07.2025 00:34 — 👍 0 🔁 0 💬 0 📌 0
Finally found this in Pittsburgh!!! This is so good!😁
02.07.2025 01:32 — 👍 1 🔁 0 💬 0 📌 0
This was so heartbreaking but beautiful!
27.06.2025 02:44 — 👍 0 🔁 0 💬 0 📌 0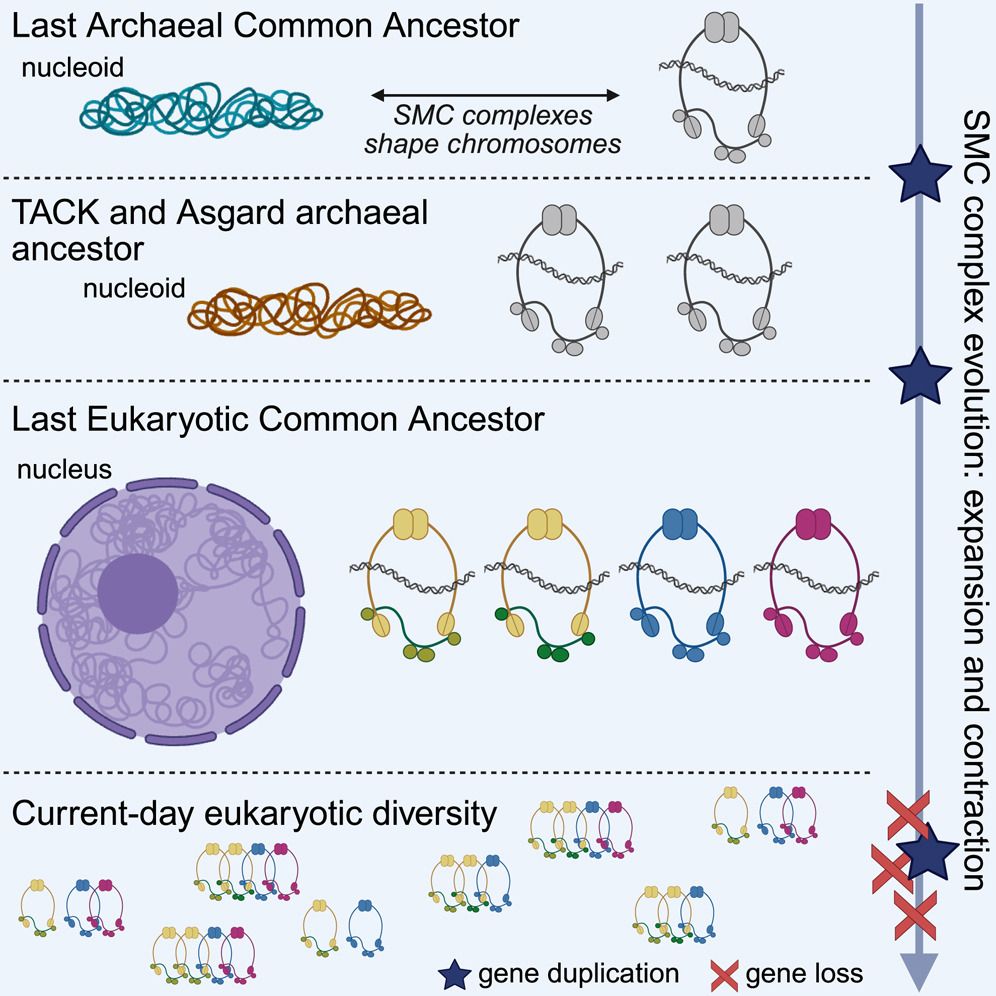
Excited to share our new paper in @cellreports.bsky.social that reshapes our understanding of chromosome organization's deep evolutionary roots! Our work dives into the origins of the machinery that structures our very genomes.
🔗: doi.org/10.1016/j.ce...
#Genomics #Evolution #CellBiology #LECA
FN is taking a summer vacation 🏖️, but we'll be back in September with more exciting talks on chromatin and transcription!
If you've got a new story you'd like to share with the FN audience this fall, fill out the form here: docs.google.com/forms/d/e/1F...