SOOOO MANY GENOMICS MODELSSSS! 😱 Often unclear which is best since they benchmark differently! In this preprint, we introduce GAME, a new framework that utilizes APIs to enable sustainable, uniform model evaluation so we can see which is actually best for each task. doi.org/10.1101/2025...
11.07.2025 07:33 — 👍 20 🔁 9 💬 1 📌 1

Black and white photos of a thylacine (now extinct) and a grey wolf, in the same pose, looking pretty similar!

Lateral adult skull of the c. thylacine and d. wolf. Ventral adult cranium of the e. thylacine and f. wolf. They also look pretty similar!
We tile through ~300 ultra conserved elements 10bp at a time in 6 taxa (some alive, some extinct), to ask, are they the reason the two skulls below- separated by 160 million years of evolution - are so similar?
(image from former PhD student Laura Cook)
11.07.2025 21:46 — 👍 4 🔁 2 💬 1 📌 0
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
13.06.2025 04:30 — 👍 164 🔁 49 💬 11 📌 10

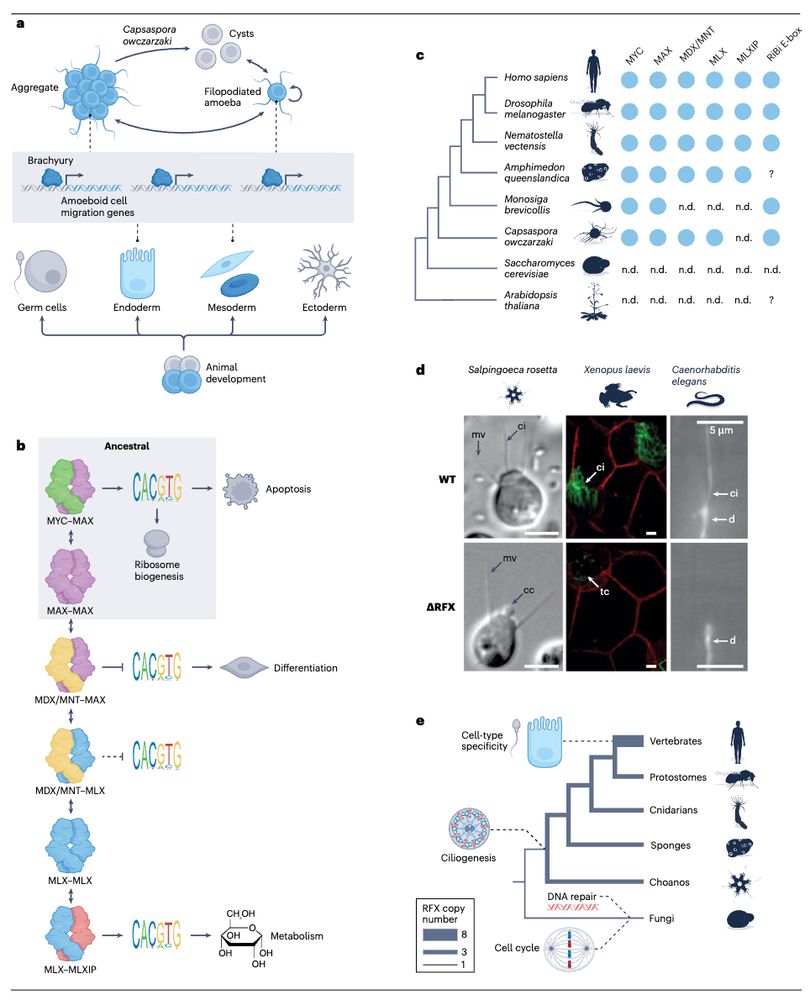
Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
27.05.2025 12:19 — 👍 245 🔁 110 💬 7 📌 9
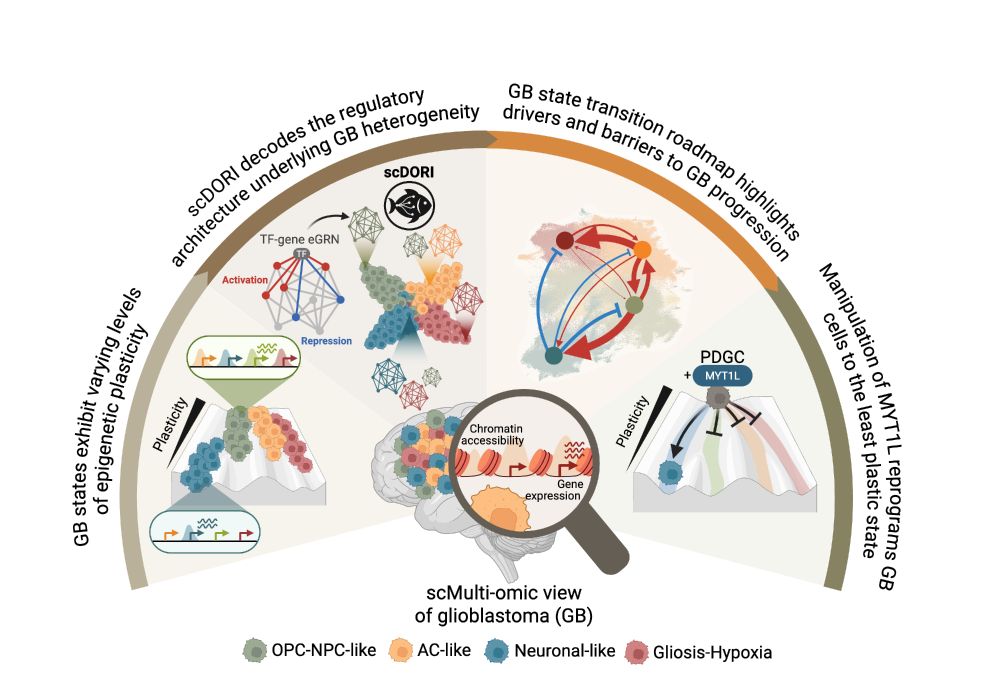
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with @bayraktarlab.bsky.social @oliverstegle.bsky.social and @moritzmall.bsky.social, Preprint at end🧵👇
16.05.2025 10:04 — 👍 76 🔁 29 💬 1 📌 6
Join us on Wednesday next week for two exciting talks on transcription regulation from @kasitc.bsky.social and @davidsuter.bsky.social!
You can register at: us06web.zoom.us/webinar/regi...
15.05.2025 18:35 — 👍 17 🔁 10 💬 0 📌 3
Very happy to share the peer-reviewed version of our paper in which we study the formation and function of pair-wise and multi-way enhancer-promoter interactions in gene regulation (see thread below): www.nature.com/articles/s41...
13.05.2025 09:41 — 👍 102 🔁 33 💬 3 📌 3
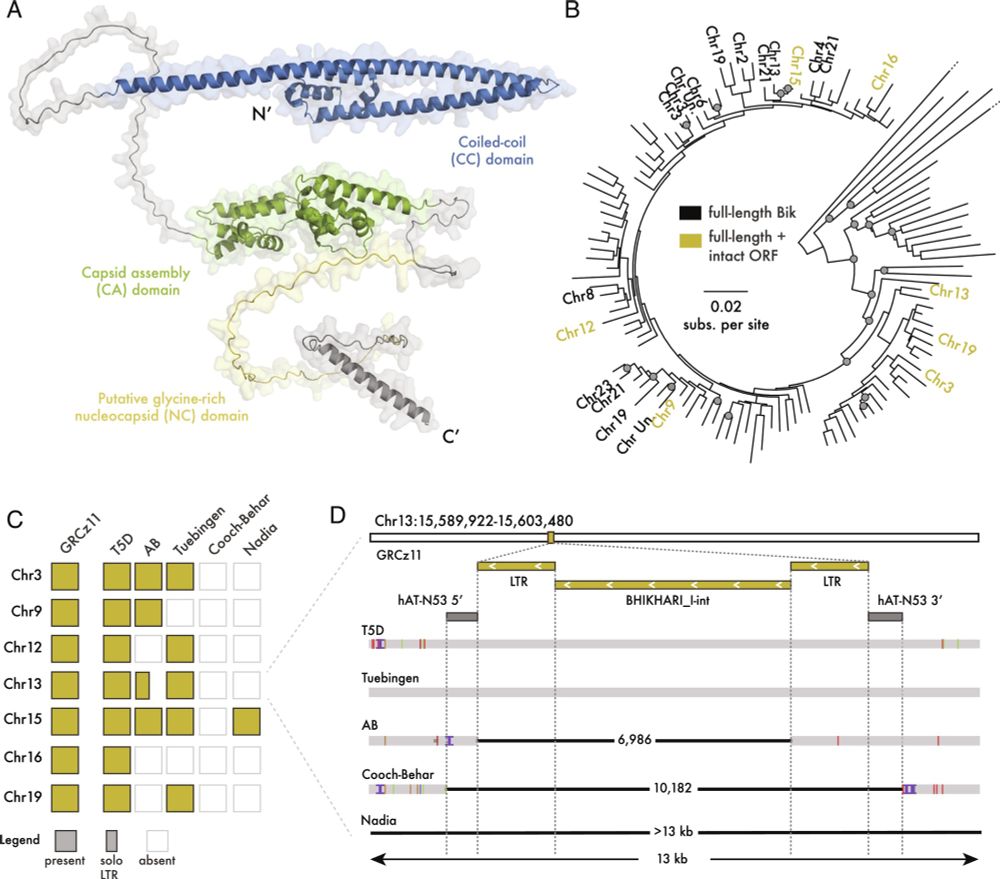
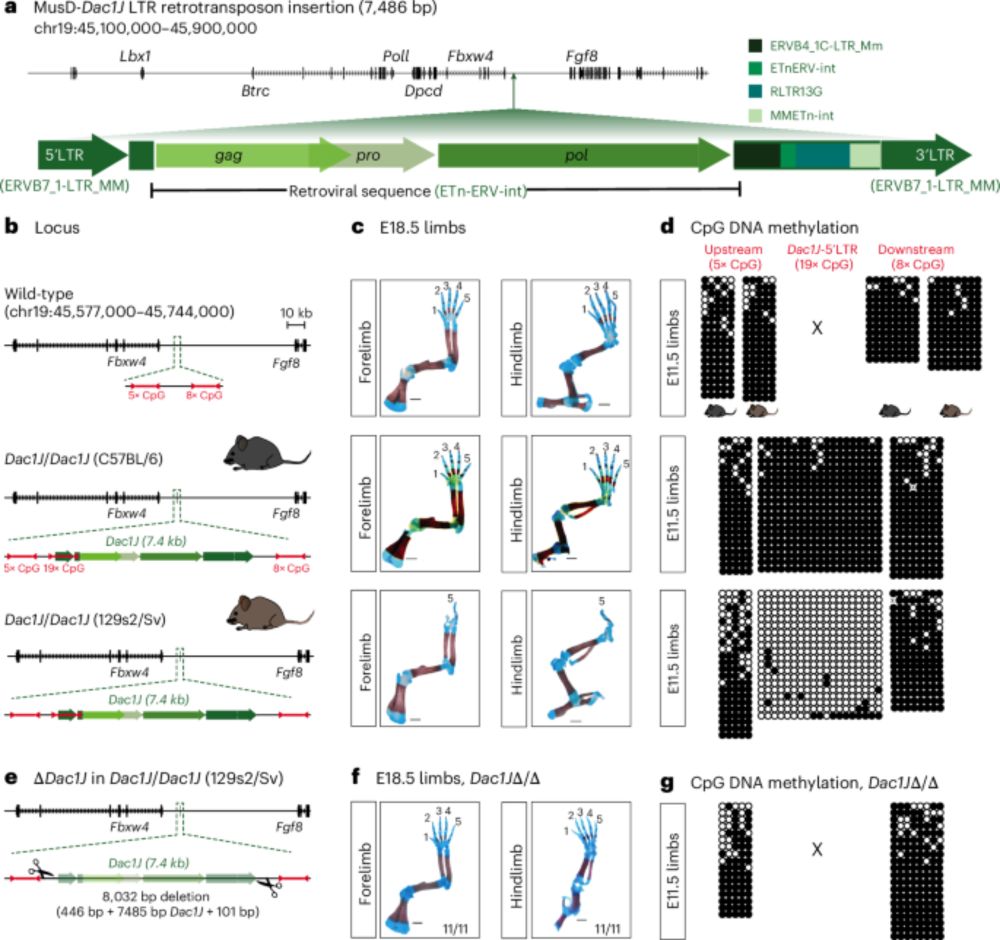
Gag proteins encoded by endogenous retroviruses are required for zebrafish development | PNAS
Transposable elements (TEs) make up the bulk of eukaryotic genomes and examples abound
of TE-derived sequences repurposed for organismal function. ...
💥🥳 At long last, our latest paper is out!
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
30.04.2025 10:44 — 👍 258 🔁 108 💬 14 📌 21
🎉 This paper has been a long time and a labour of love (and hardship) for multiple group members, but, finally: we MPRA'ed 25k introgressed variants (Denisovan and Neanderthal) segregating at allele frequencies > 0.15 in humans today to evaluate their potential to regulate gene expression.
05.05.2025 02:43 — 👍 83 🔁 29 💬 5 📌 0

Programmatic design and editing of cis-regulatory elements
The development of modern genome editing tools has enabled researchers to make such edits with high precision but has left unsolved the problem of designing these edits. As a solution, we propose Ledi...
Our preprint on designing and editing cis-regulatory elements using Ledidi is out! Ledidi turns *any* ML model (or set of models) into a designer of edits to DNA sequences that induce desired characteristics.
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
24.04.2025 12:59 — 👍 115 🔁 37 💬 2 📌 3
MPRAbase a Massively Parallel Reporter Assay database
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
One of the toughest parts of the field of massively parallel reporter assays to measure >~thousands of elements is that there are hundreds of pubs using them, but no central repo to easily locate the results....until now! Great collab w/ Jingjing Zhao, Ilias G-S, and @nadavahituv.bsky.social!!
22.04.2025 18:52 — 👍 24 🔁 9 💬 0 📌 0
Vidéos of the symposium are now available online @collegedefrance.bsky.social Just click and enjoy!🙏🏼🤘
18.04.2025 10:04 — 👍 14 🔁 5 💬 0 📌 0
I am elated to share that our manuscript describing Variant-EFFECTS, a high-throughput technology we developed to precisely and quantitatively measure the effects of CRISPR-mediated edits on gene expression, is now published at @cellpress.bsky.social: authors.elsevier.com/c/1kxgiL7PXu...
17.04.2025 18:19 — 👍 96 🔁 35 💬 4 📌 4
Qualtrics Survey | Qualtrics Experience Management
The most powerful, simple and trusted way to gather experience data. Start your journey to experience management and try a free account today.
Join the UBC and SFU Postdocs for a social at the Bloedel Conservatory and Queen Elizabeth Park on Thursday 17th April!
4-5 PM - Bloedel Conservatory (Free Entry)
5 PM - BBQ at QE Park
Feel free to join for both parts of the event or just one.
Register now: ubc.ca1.qualtrics.com/jfe/form/SV_...
08.04.2025 21:03 — 👍 1 🔁 4 💬 0 📌 0
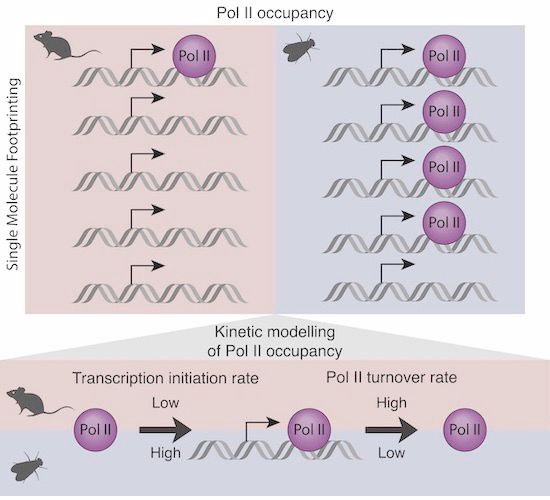
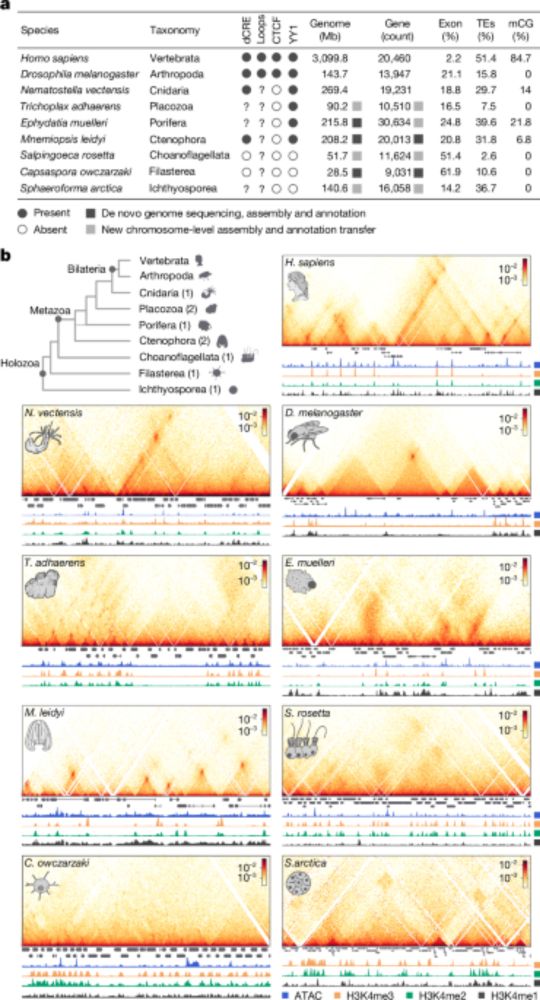
The importance of moving away from bulk! Occupancy of Pol II at promoters is dramatically different between fly and mouse cells! When looking single molecule! Proud of the team! @kasitc.bsky.social @molinalab.bsky.social doi.org/10.1038/s443...
02.04.2025 08:47 — 👍 115 🔁 37 💬 2 📌 1

Celebrating 10 years of our lab with a new preprint:
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
29.03.2025 12:45 — 👍 149 🔁 58 💬 6 📌 2
Join us for the first edition of the Cascadia Advanced Genomic Technologies meeting! We aim at fostering collaborations to tackle critical challenges in mammalian genomics. Info and registration link below 👇
26.03.2025 17:01 — 👍 2 🔁 0 💬 0 📌 0
CAGT: Cascadia Advanced Genomic Technologies
June 19-20 2025, Vancouver, BC, Canada
Join us June 19-20, 2025 in Vancouver BC for the Cascadia Advanced Genomic Technologies meeting! Featuring Keynote speaker Calin Plesa @calin.bsky.social Abstract deadline April 30🔔, but registration is capped so don't wait! 😱
de-boer-lab.github.io/CAGT_meeting/
26.03.2025 16:47 — 👍 5 🔁 5 💬 1 📌 1
A though provocative read genome.cshlp.org/content/35/3.... While estrogen response triggers transcriptional and 3D chromatin changes, only a select few structural shifts are likely to subtly contribute to transcriptional regulation 🤔
19.03.2025 16:37 — 👍 1 🔁 1 💬 1 📌 0

In this study, Huang et al. demonstrate how a single enhancer independently controls the expression of two distal and functionally unrelated genes.
Learn more here:
➡️ tinyurl.com/gd352235
14.03.2025 15:10 — 👍 17 🔁 6 💬 0 📌 1
je suis salarié de l'ina où j'écris pour la revue des médias, et je me spécialise dans les sujets aux intersections du journalisme et de l'intelligence artificielle
xeutrope@ina.fr
Crédit photo de profil : Radio France / Christophe Abramowitz
Journaliste pour la cellule enquêtes de @telerama.bsky.social. Essayiste anxieux. "Apocalypse Nerds" chez @divergences.bsky.social (19/09).
☎️ Signal : tesq.37
https://linktr.ee/oliviertesquet
Journaliste.
Co-responsable du pôle « Enquête » de Mediapart.
Auteur de livres : « La Troisième Vie », « D’Argent et de Sang », « Pas tirés d’affaires », « Avec les compliments du guide »…
www.mediapart.fr
Observatoire des médias depuis 1996.
https://www.acrimed.org
S'inscrire à l'infolettre d'Acrimed : https://www.acrimed.org/L-infolettre-d-Acrimed
Professeur, Auteur, Streamer.
📺 Sur http://twitch.tv/clemovitch dans Le Café Rhétorique (lundi, mercredi, vendredi)
🎭 Sur scène avec « L’art de ne pas dire »
🎫 https://clemovitch.com/
📩 clemovitch@20ty.gg
Rhinocéros - analyse des médias chez Blast
Ça ira - émission pour L'Huma
Insta: lumi_sco
Même bio que sur Copains d'Avant
Islamopatissier, cofondateur de Gandi, auteur de http://confessions-voleur.net. Porteur du projet http://caliopen.org (contact: laurent @ http://brainstorm.fr)
Le gars des chroniques là, dans la presse en ligne indépendante tu vois, si, tu sais, le 3615, mes chers compatriotes et tout
Gardienne d'Horizon
Juger un peu moins, comprendre un peu plus, créer davantage
https://hacking-social.com
http://youtube.com/c/horizongull/
Hacktivist, ex-Mozilla, podcaster (l"Octet Vert", in French). Thinks that bicycles are a good chunk of the solution to the climate problem.
Legal stuff, mostly about privacy. Former legal and public affairs @ Clever Cloud 🌥️ and Qwant // founder of Numerama 📰 🔍 // fcnantes supporter 🐥// chess player ♟️
Le magazine scientifique d'actualité
Pour vous abonner : https://www.epsiloon.com/
Nos derniers articles en libre accès : https://linktr.ee/epsiloon
Journaliste au service #Planète au Monde. Rubrique #climat, COP, mobilisations, réchauffement et apocalypses diverses. Mes tweets sont en poils de yak.
Médecin de formation, #journaliste médico-scientifique par vocation. #Freelance
✍ Nouvelle adresse du #blog 'Réalités Biomédicales' https://www.lemonde.fr/realites-biomedicales/
✍ Archives (734 billets) https://www.lemonde.fr/blog/realitesbiomedicales/
CEO of AgroClimat2050 | PhD in agrometeorology | Vice-President & Stormchaser Infoclimat |
Conference / Speaker #agriculture #globalwarming
Chercheur, vulgarisateur et youtubeur sur https://www.youtube.com/@Fouloscopie
Compte officiel de la chaîne Youtube "Science étonnante"
Scientist, Sci-Fi Author, and Tech Nerd wandering the world. Obsessed with #biotech, #AI, and frontier tech.
Join me @ connectedideasproject.com for thoughts on tech, policy, and life.
Maison d'édition indépendante créée en 1996, Le Bélial’ consacre son catalogue aux littératures de genre (SF, fantasy fantastique) et édite la revue Bifrost.
🚀🐉👻
Bannière : Amir Zand
www.belial.fr