
The goal is to provide a lot of flexibility in a familiar framework, while delivering robust analysis methods coupled with extensive visualization options.
Check it out!
github.com/lrylaarsdam/...
@acadey.bsky.social
Professor of Molecular & Medical Genetics at OHSU. Runner.

The goal is to provide a lot of flexibility in a familiar framework, while delivering robust analysis methods coupled with extensive visualization options.
Check it out!
github.com/lrylaarsdam/...
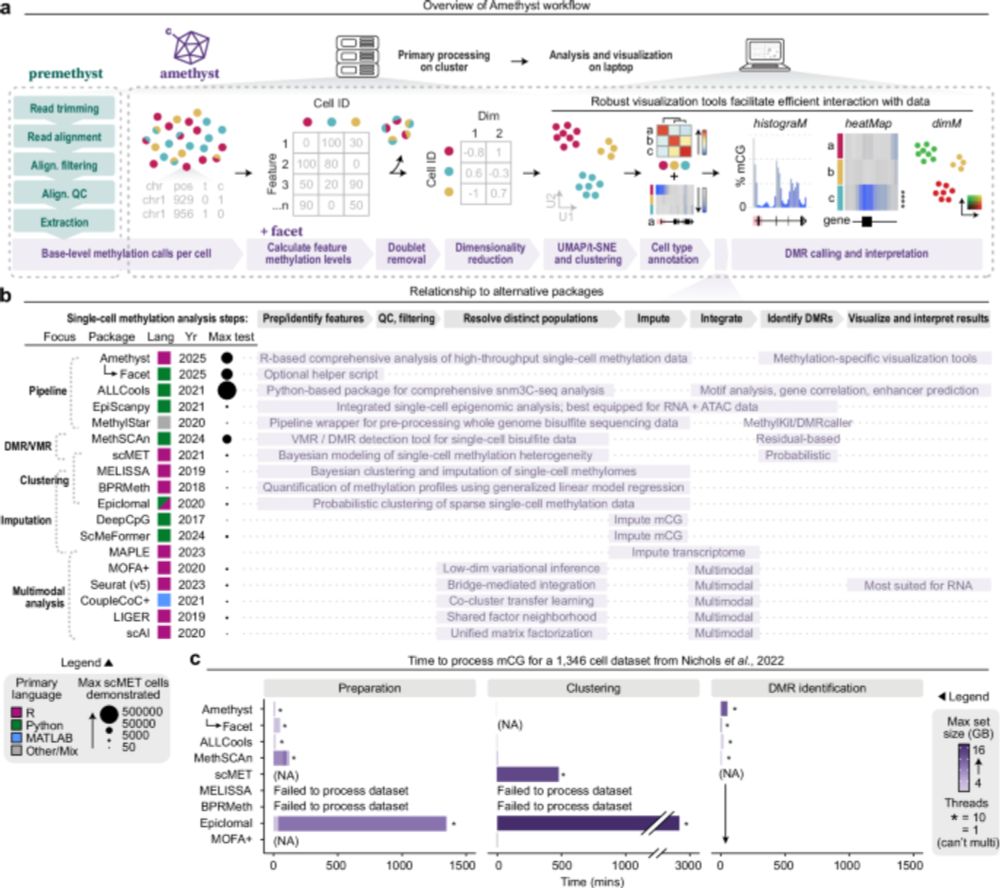
Really excited to have our single-cell DNA Methylation analysis tool, Amethyst, published! Led by Lauren Rylaarsdam, who developed the tool as a way to tackle her own projects and developed it into a comprehensive scDNAm analysis package.
www.nature.com/articles/s42...


Super excited about first Shendure/Baker Lab collaboration & preprint on a multiplex sequencing-based strategy for screening de novo proteome editors in mammalian cells. Kudos to the brilliant Chase Suiter (not here) & @greenahn.bsky.social on the work! Preprint here:
www.biorxiv.org/content/10.1...
Awesome!
01.10.2025 18:53 — 👍 1 🔁 0 💬 0 📌 0
Huge thanks to all co-authors and the inimitable @jbuenrostro.bsky.social for supporting this new direction - I got to think about evolution, engineering, chromatin biophysics, and modeling during the course of this project. For more, check out the preprint: www.biorxiv.org/content/10.1... 8/9
11.09.2025 19:55 — 👍 19 🔁 4 💬 2 📌 0Excited to share my postdoc work, out on bioRxiv today! Histones package DNA into nucleosomes to form the building blocks of chromatin, but how modular and programmable is this system? 1/9
11.09.2025 19:55 — 👍 85 🔁 32 💬 1 📌 2I am excited to work with 10x to take single-cell technologies to the next level - lots of new directions!
07.08.2025 20:44 — 👍 0 🔁 0 💬 0 📌 0
We have created a new DNase I- & ATAC-seq peak caller that uses an adaptive background model that controls for copy number variation & aneuploidy. It performs a per-nucleotide test (+FDR correction) and is very fast. Please try it out and give us feedback!
github.com/vierstralab/...
Thanks for this! What is your take when sample quality means only modest ATAC-seq quality is possible. (too low is not usable, so let's say borderline) In many cases, eg primary tumors etc... high quality can never be achieved. What is the threshold for different analyses that can be performed?
14.05.2025 16:23 — 👍 2 🔁 0 💬 0 📌 0Happy to share our new review on molecular circuits for genomic recording with @chenomics.bsky.social! Here, we present our views on the remaining challenges in the field of genomic recording for reconstructing cell-fate decisions. (1/2)
06.05.2025 14:40 — 👍 16 🔁 5 💬 1 📌 0
Cuts to #NIH funding don't just hurt scientists, but everyone who depends on new treatments, cures, & medical breakthroughs. Research can't thrive without strong, sustained support. Read more in this @60minutes.bsky.social segment featuring former NIH director Francis Collins: buff.ly/REjVWPc
29.04.2025 20:02 — 👍 17 🔁 17 💬 0 📌 0Delighted to see this out! Fun fact: I actually started my lab @ UCSF w/ goal of understanding tRNA txn regulation, which aligned perfectly w/ @genophoria.bsky.social & Sohail! 6+ years of amazing collaboration & stellar work by Siyu, Albertas, & co & we're making real progress w/ more to come!
01.05.2025 03:22 — 👍 21 🔁 4 💬 0 📌 0
Want to work with us on DNA methylation and rare genetic disease? Fully funded PhD project with deadline 16th May:
www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics
Whenever my kid sees a Cybertruck she says "that's a stupid truck" and I feel like I'm succeeding at parenting
20.04.2025 23:36 — 👍 166 🔁 4 💬 7 📌 0
2) HyDrop-v2: with a new bead design it provides scalable and cost-effective generation of scATAC-seq atlases. With HyDrop atlases of the fly embryo and mouse cortex we show that CREsted models trained on HyDrop data are equivalent to models trained 10x atlases. www.biorxiv.org/content/10.1...
04.04.2025 09:04 — 👍 14 🔁 5 💬 1 📌 0
Very proud of two new preprints from the lab:
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
Proud to see so many fellow scientists and citizens standing up for science today! Every treatment option I'm able to offer my patients came to be because of careful persistent scientific inquiry. We cannot afford to be shortsighted when it comes to biomedical research. #StandUpForScience
08.03.2025 01:03 — 👍 4 🔁 1 💬 0 📌 0Interesting work from the Tomkova group relating DNA hypermethylation of polycomb-repressed regions with gene upregulation. #Epigenetics #DNA_Methylation
14.02.2025 13:41 — 👍 9 🔁 5 💬 1 📌 0What's happening is precisely outlined in Project 2025. Read it to understand what's coming next. None of this is a surprise.
Also, I know this is extremely unnerving if u weren't expecting this. But let's not panic. Let's organize & give this the best fking fight of our lives.
"Single" cell... no editing typos on bluesky I guess.
24.12.2024 17:39 — 👍 0 🔁 0 💬 0 📌 0Finally, we leverage stepwise, indexed tagmentation to produce ATAC and DNA methylation profiles from the same cells. We believe this will be particularly useful for bridging to ATAC atlases to better facilitate Cell type annotation.
24.12.2024 17:36 — 👍 0 🔁 0 💬 0 📌 0To enable more sequencing options, we also developed primers for direct preparation for sequencing on Ultima Genomics instruments. We sequenced >140k cells from human cortex.
24.12.2024 17:36 — 👍 0 🔁 0 💬 1 📌 0We also show you can use enzymatic conversion methods instead of bisulfite.
24.12.2024 17:36 — 👍 0 🔁 0 💬 1 📌 0
The core of the improvement is an additional layer of indexing. This is done while maintaining the same molecular structure as sciMETv2 which enables compatibility with our sciMET-CAP capture workflow. (genomebiology.biomedcentral.com/articles/10....)
24.12.2024 17:36 — 👍 0 🔁 0 💬 1 📌 0
Excited to have our sciMETv3 paper out today in Cell Genomics! We can produce hundreds of thousands of sibgle-cell DNA methylome libraries in a single experiment!
(Plus lots more)
www.cell.com/cell-genomic...
Just joined. Not sure how much I will post or follow. I have enjoyed the post-Twitter zen, but I do miss the collation of useful info and papers in the field.
16.12.2024 17:46 — 👍 0 🔁 0 💬 0 📌 0