🎉 Congrats to MSK postdoc research scholar Dr. Brendan Camellato for being named a 2025 @jcchildsfund.bsky.social Research Fellow! His work will focus on how cell signaling drives #stemcell differentiation with single-cell resolution. #JCCFellow
24.06.2025 17:13 — 👍 7 🔁 3 💬 0 📌 0
Congrats Brendan Camellato for his Jane Coffin Childs Fellowship!! 🎉🎉
24.06.2025 22:05 — 👍 9 🔁 1 💬 0 📌 0
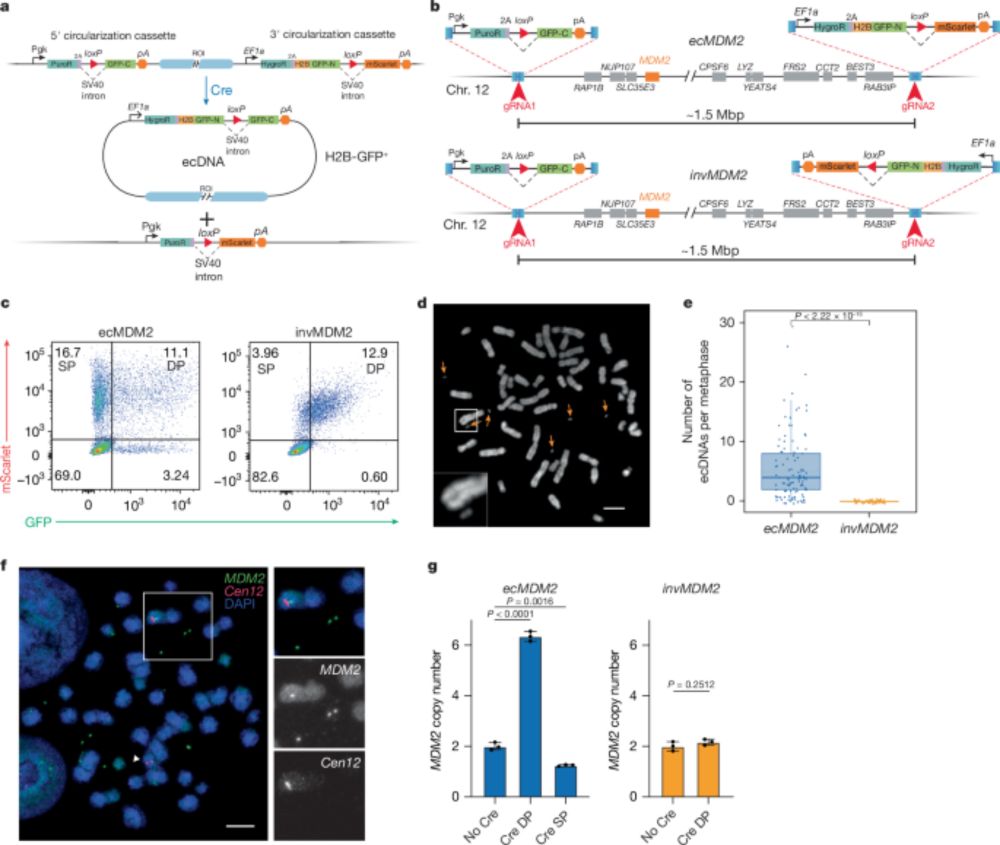
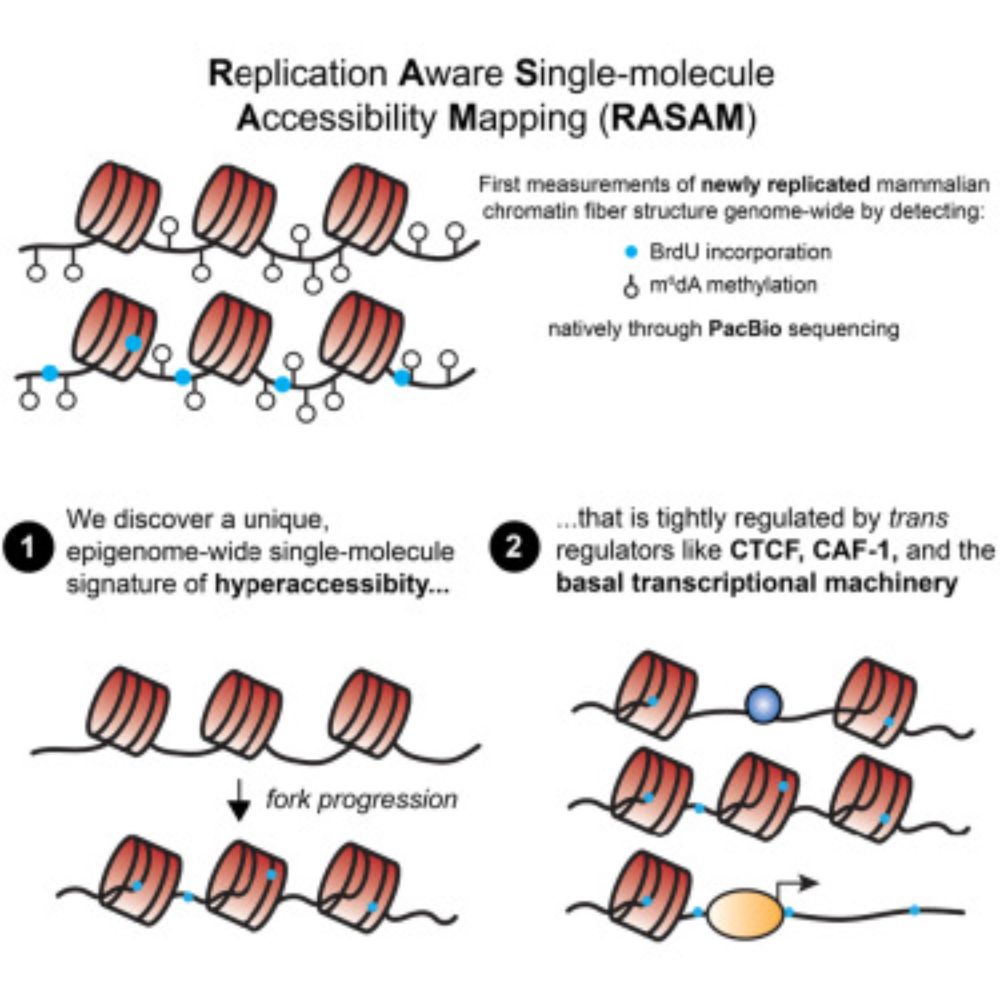
We're thrilled to share the published version of our DRT9 story, online today @nature.com! Congratulations to all authors!
www.nature.com/articles/s41...
28.05.2025 21:06 — 👍 56 🔁 18 💬 0 📌 1
Thank you, Emma!!
28.05.2025 00:28 — 👍 1 🔁 0 💬 0 📌 0
We’re still thinking about what this means for actual embryonic development, but it’s exciting to observe such effects in a tractable stem-cell-derived model! Huge thanks to everyone in the @jshendure.bsky.social lab—especially Jay—for supporting this project from start to finish! (13/13)
27.05.2025 14:56 — 👍 0 🔁 0 💬 0 📌 0

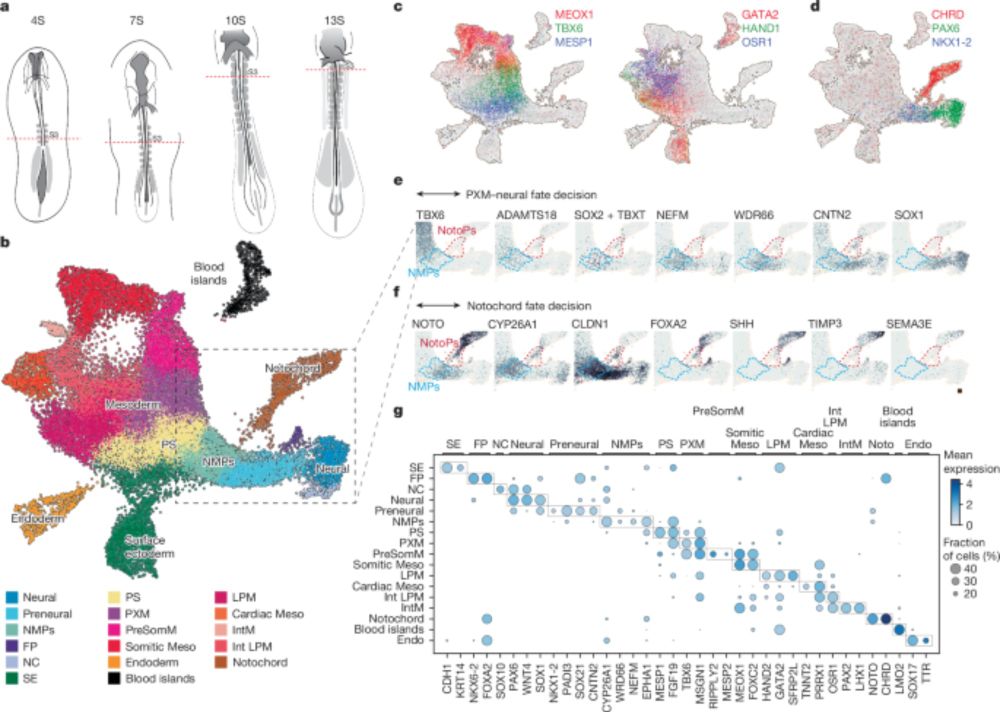
Part of Image5 in the preprint, describing how major clades share similar cell-type compositions and cell numbers within each gastruloids.
Even after resetting spatial context, we saw persistent clonal biases—suggesting an intrinsic, heritable factor shaping fate decisions, independent of spatial cues like Wnt signaling. (11/13)
27.05.2025 14:56 — 👍 1 🔁 0 💬 1 📌 0
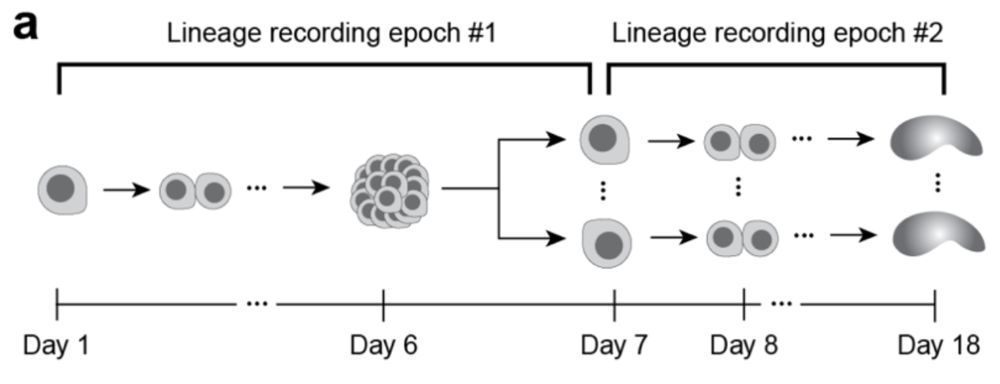
Image describing two-epoch experiment, where we first start with single cell that makes a cell colony, which is dissociated in the second epoch and each made into a monoclonal gastruloid.
In this “tree-of-trees” experiment, we dissociated a monoclonal colony into single cells (resetting spatial context) and used them to generate separate gastruloids. (10/13)
27.05.2025 14:56 — 👍 0 🔁 0 💬 1 📌 0

Image showing early clonal bias from first, third and 12th cell division influencing cell fate decision between somitic vs. neural cell fate.
Together, we found: Cell fates can diverge as early as the first cell division! Sister cells can follow drastically different paths—e.g., one lineage becomes mostly somites, another mostly neurons! (8/13)
27.05.2025 14:56 — 👍 0 🔁 0 💬 1 📌 0

Image showing the lineage tree of monoclonal gastruloid.
Once we knew monoclonal lineage recording worked, we teamed up with @cxqiu.bsky.social—an amazing and generous computational biologist—to dive into the lineage trees. (7/13)
27.05.2025 14:56 — 👍 1 🔁 0 💬 1 📌 0
(Side note: Sam also used this approach to generate monoclonal embryoid bodies for perturbation screening with @sdomcke.bsky.social and @cxqiu.bsky.social.) bsky.app/profile/sdom... (6/13)
27.05.2025 14:56 — 👍 1 🔁 0 💬 1 📌 0

Image describing the monoclonal gastruloid induction protocol.
To solve this, Sam developed a robust protocol to create “monoclonal gastruloids”—gastruloids seeded from a single cell. These were ideal for DNA Typewriter lineage reconstruction! (5/13)
27.05.2025 14:56 — 👍 0 🔁 0 💬 1 📌 0

Image describing conventional gastruloid induction protocol, which starts with 300-500 cells.
However, conventional gastruloids start with 300–500 cells, and that’s a problem for us—lineage tracing from 500 cells gives you 500 independent trees, masking clonal info from earlier developmental events. (4/13)
27.05.2025 14:56 — 👍 1 🔁 0 💬 1 📌 0

Image of 3D gastruloids, highlighting their morphological development and cell-type differentiations
Mouse 3D gastruloids seemed like the perfect testbed—they mimic key features of early mammalian development in both shape and cell types. (Image from Turner @gastruloids.bsky.social & Martinez Arias @amartinezarias.bsky.social : doi.org/10.1002/bies...) (3/13)
27.05.2025 14:56 — 👍 1 🔁 0 💬 1 📌 0
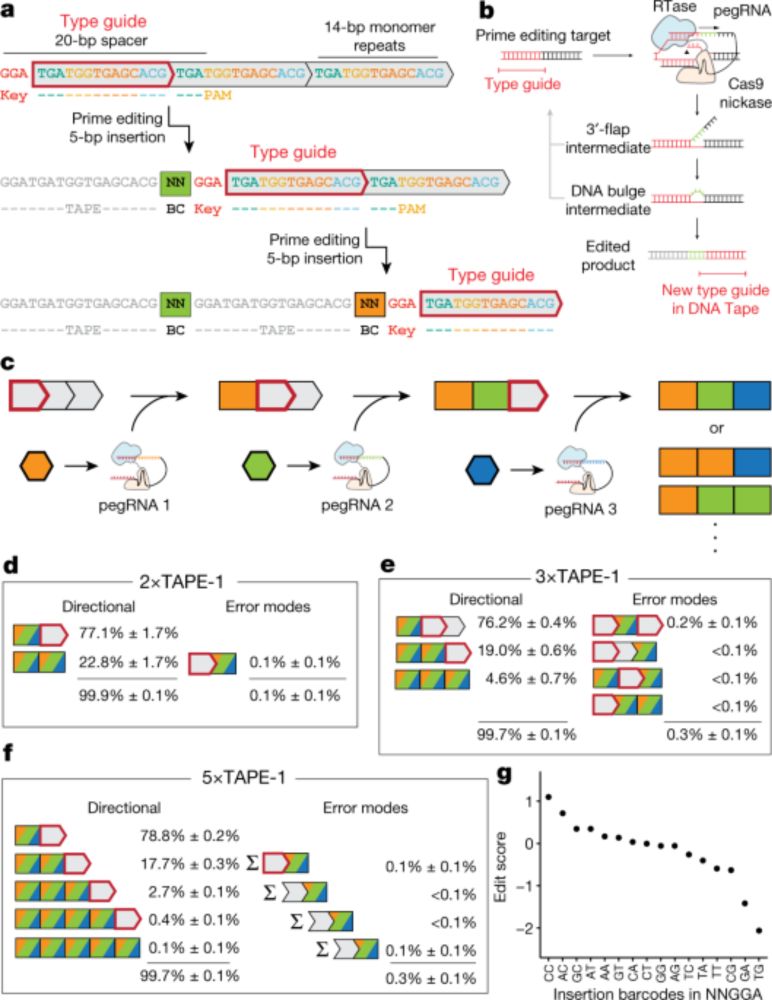
A time-resolved, multi-symbol molecular recorder via sequential genome editing - Nature
A DNA memory device, DNA Typewriter, uses sequential prime editing to record the order of multiple cellular events.
We previously showed that DNA Typewriter can record high-resolution lineage trees in cultured cells (HEK293T). www.nature.com/articles/s41... But could we use it to study how clonal memory shapes cell fate decisions during development? (2/13)
27.05.2025 14:56 — 👍 1 🔁 1 💬 1 📌 0
Thank you for kind words!!
07.05.2025 12:41 — 👍 1 🔁 0 💬 0 📌 0
Thanks, Eric!
07.05.2025 12:40 — 👍 0 🔁 0 💬 0 📌 0
We are very thankful to Jay Shendure @jshendure.bsky.social and the lab for fostering our views that led to this review, @cp-trendsgenetics.bsky.social for all their help, and the funding agencies (NHGRI, NCI, and @damonrunyon.org) that made this work (and our previous works) possible! (2/2)
06.05.2025 14:40 — 👍 2 🔁 0 💬 0 📌 0
Happy to share our new review on molecular circuits for genomic recording with @chenomics.bsky.social! Here, we present our views on the remaining challenges in the field of genomic recording for reconstructing cell-fate decisions. (1/2)
06.05.2025 14:40 — 👍 15 🔁 5 💬 1 📌 0

a baby is eating a piece of food while sitting on the ground .
ALT: a baby is eating a piece of food while sitting on the ground .
Trying a slightly new model (for us at least) of #openscience. With @ronanchaligne.bsky.social and @karolisk.bsky.social, we pitted 10x Genomics Chromium GEM-X against Illumina PIPseq V head-to-head to understand these two platforms for single-cell genomics. Some thoughts off the bleeding edge: /1
14.03.2025 13:29 — 👍 60 🔁 18 💬 1 📌 3
New year, new beginnings! 🎉
The Aviram Lab will officially open its doors on March 1st @mskcancercenter.bsky.social!
We’ll study microbial immune systems and their connection to fundamental cellular processes like #RNA transcription and #DNA integrity.
#NewPI #AcademicSky #WomenInSTEM
(1/2)
02.01.2025 16:01 — 👍 69 🔁 18 💬 7 📌 1

‘DNA Typewriters’ Can Record a Cell’s History
Labs around the world are trying to turn cells into autobiographers, tracking their own development from embryos to adults.
Imagine a mouse in which all 10 billion cells keep a diary from fertilization. Imagine sentinel cells recording your experiences and sending out dispatches in the form of DNA. Here's a story I wrote about cell recorders 🧪 Gift link: nyti.ms/3Z3Mqza
25.11.2024 16:19 — 👍 109 🔁 33 💬 2 📌 9
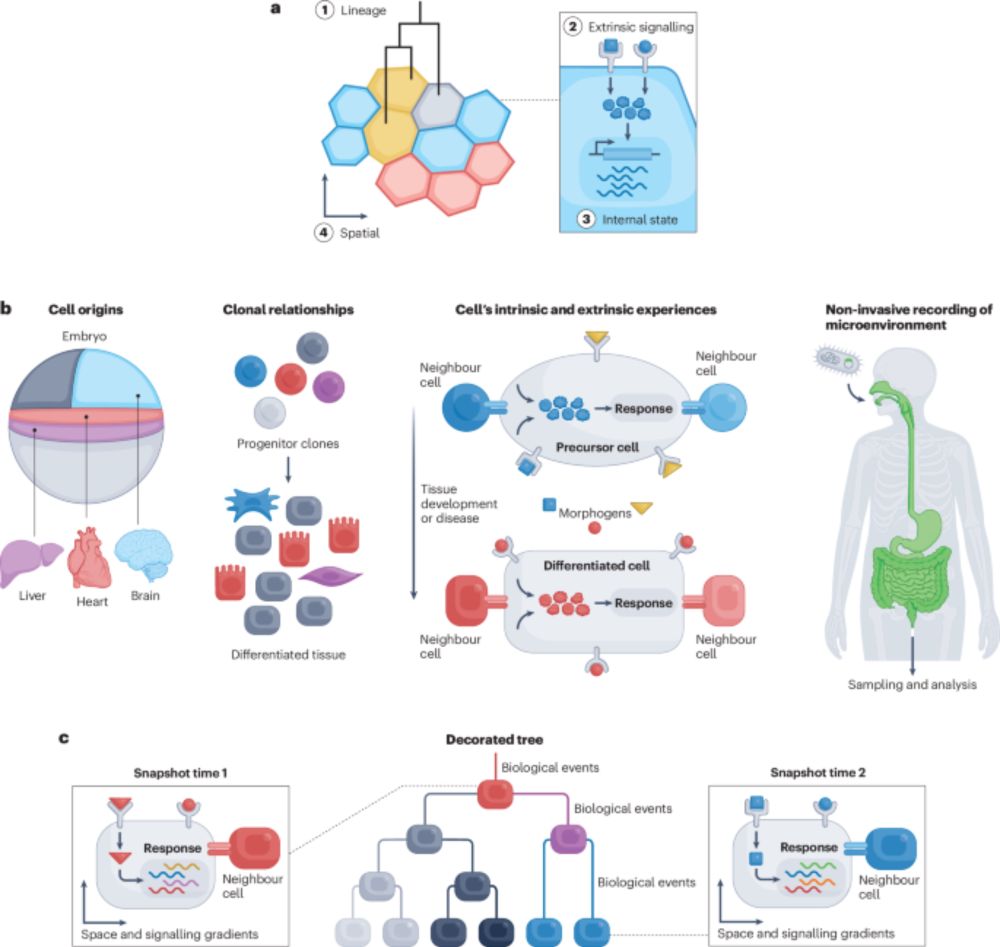
The lives of cells, recorded - Nature Reviews Genetics
Recent advances in genome engineering are enabling the recording of cellular histories into genomes, with single-cell and spatial omics technologies enabling their reconstruction into cellular lineage...
"The lives of cells, recorded"--our new review on genomic recording systems and how they can reveal the dynamics of multicellular development. A pleasure to work on this with amazing colleagues from the Allen Discovery Center for Cell Lineage Tracing.
www.nature.com/articles/s41...
25.11.2024 17:28 — 👍 144 🔁 58 💬 2 📌 4
Nobuhiko (Nobu) Hamazaki, Ph.D. Assistant prof. @University of Washington (UW). Stem cell engineering, germ cell and embryonic development.
Computational Biologits @Kelley lab @Calico. Previously @UW CSE and GS @Shendure lab.
PhD Candidate @CRG.eu with @larsplus.bsky.social | Computational Biology & Single Cell Epigenetics
Postdoc at Memorial Sloan Kettering Cancer Center, New York | Fascinated by cancer cell plasticity, tumor microenvironment, immunology and therapy resistance
Postdoc at Sloan Kettering Institute in http://sfeirlab.com | former Kapoor Laboratory at RU | interested in #CancerBiology #DNARepair #DrugResistance #Mitosis | opinions my own | she/her
Tri-I MD-PhD | Ganesh Lab |
Postdoc at the Vierbuchen lab (MSKCC), previously PhD at Institut Imagine (Cantagrel/Colleaux lab) /// I do mouse brain organoids to characterize early development and understand what makes each brain unique
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
Molecular & structural biologist CNRS with a big respect for Transcription Factors, RNA Pol II & Mediator
#CryoEM #ESR
Advocating for #basicresearch #womeninSTEM #preprint
Message in English or French.
https://bsi-lille.cnrs.fr
#DevBio Lab at NorthwesternU Studying Pluripotency, Neural Crest Stem Cells and the Evolution of Vertebrates. Recovering Former Dept. Chair, Diversity Advocate. Past President Society for Developmental Biology.
Professor at KTH, NY Genome Center, SciLifeLab, working on functional genomics and human genetics.
Chromatin and cryoEM afficionada. Still a fan of crystallography. Avid Colorado hiker. Will call out ugly nucleosome cartoons. Come for the science, stay for the mountain pictures and snark. Opinions and snark are my own.
Assistant Professor of Cell Biology at Harvard Medical School | HHMI Freeman Hrabowski Scholar | Nexus of chromatin, transcription, replication, and epigenetics. farnunglab.com
Professor and Head of @WUSM_BMB. Studying protein PTMs in chromatin biology. #TeamMassSpec cheerleader. Working towards changing academia. Views my own 🇺🇸 🇲🇽
Biologist. Chromosomes | dinoflagellates | birds. Happy in academia! Umass Chan Medical School, HHMI
http://www.dekkerlab.org
Genomics, AI, sequence-to-function models, mechanisms of the cis-regulatory code. Investigator at the Stowers Institute.
Head of Genome Biology Dept @EMBL,
Scientist, Principal investigator, Professor
Exploring genome regulation during development,
and everything to do with enhancers.
3D genome, chromatin topology,
cell_fate, embryonic Development,
Single Cell genomics
PI @ UW Genome Sciences | chromosome biology | 3D genome | chromatin | single cell & spatial biology
We develop technologies to study genomes and the things that are made from them. Avid FISHer.
beliveau.io
Laboratory for Chromatin and Spatial Neurobiology
We work at the intersection of chromatin, synapses, and neurobiology to understand how the brain stores memory over long timescales.
creminslab.com
x.com/creminslab
https://videocast.nih.gov/watch=55006