Long in the making, but happy to present the Chlamydomonas chlororibosome!
Cryo-ET🔬reveals a large new domain on the small subunit, built from multiple extensions in conserved ribosomal proteins.
bioRxiv 📖: shorturl.at/q44tG
This suggests greater chlororibosome diversity than expected!
1/n 🧵
10.02.2026 08:35 — 👍 87 🔁 29 💬 2 📌 5
Hot Topic Conference
Support for an "Eduard Strasburger Hot Topic Conference" for DBG's members
For #ECR’s:
DBG’s Hot Tpoic Workshop
on Cryo-Electron Tomography + FIB-milling
13 – 15 April 2026
In @cellarchlab.com 's lab @biozentrum.unibas.ch
Organised by @tamb-o.bsky.social, Karen Zinzius, @fannyleblanc.bsky.social
Details: www.deutsche-botanische-gesellschaft.de/en/about-us/...
03.02.2026 17:24 — 👍 11 🔁 9 💬 0 📌 0
You like #LLPS? #TeamTomo? #Rubisco? Check out our latest preprint!
What a great collaboration this has been!
30.01.2026 15:38 — 👍 30 🔁 11 💬 0 📌 0
Excited to share this detailed biophysical study of #PhaseSeparation in the Pyrenoid, an algal microcompartment that fixes a third of the Earth's #CO2 🌏. We combine methods across scales, in vitro 🧪 and in vivo 🦠
Thanks for the very fun collaboration! 🤗
#PlantScience 🌾 #TeamTomo 🔬 #LivingPhysics 🧬🧶
30.01.2026 15:50 — 👍 63 🔁 18 💬 1 📌 1
Check out this cool thread on the latest preprint by Gaurav Kumar, @james-r-barrett.bsky.social, @phaips.vd.st from our lab and colleagues, dissecting the physics of Rubisco condensates!
You're in for some stunning #cryoET of the pyrenoid - in Chlamy, of course 😉
Congrats to all involved!
30.01.2026 15:54 — 👍 18 🔁 6 💬 0 📌 0
How does molecular valency shape condensate assembly and function? We used the CO2-fixing organelle in algae—the pyrenoid—to find out… 🧵
Preprint here!:
www.biorxiv.org/content/10.6...
@cellarchlab.com @phaips.vd.st @biologyatyork.bsky.social
#Rubisco #PhaseSeparation #Condensates #Pyrenoid
30.01.2026 15:20 — 👍 74 🔁 34 💬 1 📌 5
🔬🚨New preprint alert! 🚨🔬
We developed quantitative expansion microscopy (qExM) - a method to accurately count proteins in situ by combining expansion microscopy's improved labeling with statistical estimators borrowed from ecology
www.biorxiv.org/content/10.6...
#SuperResolution #CellBiology
20.01.2026 16:12 — 👍 44 🔁 18 💬 5 📌 1

a woman says " would you like to join us " with her eyes closed
ALT: a woman says " would you like to join us " with her eyes closed
📢 Open faculty position – Origins of Life
We have an opening in our section at the University of Geneva! 🧬🚀
SPREAD THE WORD
Apply here: jobs.unige.ch/www/wd_porta...
16.01.2026 17:49 — 👍 45 🔁 39 💬 1 📌 1

The latest issue is now online www.cell.com/molecular-ce...
08.01.2026 16:59 — 👍 19 🔁 7 💬 0 📌 2
Welcome to TomoGuide
A step-by-step Cryo-ET guide
If you want to learn how to process cryo-ET data using the Chlamy dataset don't forget to check : tomoguide.github.io
@phaips.vd.st
19.12.2025 16:45 — 👍 15 🔁 3 💬 1 📌 0

a man walking in a field with an umbrella
ALT: a man walking in a field with an umbrella
Come join us! Soon I will be advertising a postdoc vacancy in my group as part of my @erc.europa.eu AdG project 'DARK ROOTS'. Focus of the project will be on phylo- and metagenomic mining of novel prokaryotic lineages. I will soon post a link here - stay tuned, and please repost! #asgardarchaea
26.11.2025 15:39 — 👍 33 🔁 33 💬 0 📌 2

An Asgard archaeon with internal membrane compartments
Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...
07.11.2025 10:44 — 👍 387 🔁 170 💬 10 📌 22

We’re back with the next DinoSphere Online Seminar!
Join us on Nov 4th, we're hosting:
Edmée Royen (ULiège) - Symbiodinium
Nicolas dos Santos Pacheco (Cambridge) - Perkinsids
📅 Tue, Nov 4 - 4 PM CET/3 PM GMT
💻 Zoom link: sites.google.com/view/dinosphere
Please spread the word!
#protistonsky
28.10.2025 15:29 — 👍 19 🔁 16 💬 0 📌 2

Fig. 2.Confocal microscopy of N. punctiforme strains. Filaments from the indicated strains were taken from cultures grown in BG110 medium (not induced; A) or after incubation for 24 h in the presence of Anthoceros agrestis exudates (hormogonia induction; B) and visualized by confocal microscopy. Merged (2 channels) show the overlay of autofluorescence (magenta) and GFP fluorescence (green). Merged (3 channels) represent the overlay of magenta autofluorescence, bright field, and green GFP fluorescence. Hormogonia are identified as short filaments with small cells. Scale bar=20 μm.
💡🔬 TECHNICAL INNOVATION 💡🔬
Neubauer et al. developed a Nostoc reporter strain enabling high-throughput quantitative monitoring of hormogonia differentiation in cyanobacteria as a response to biotic and abiotic factors.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @peterszovenyi.bsky.social
24.09.2025 11:31 — 👍 9 🔁 7 💬 0 📌 0
🥨🔬!
23.09.2025 17:31 — 👍 3 🔁 0 💬 0 📌 0
🌱 Using ‘compelling’ methods, including #CryoET, researchers mapped spinach thylakoid membranes at single-molecule precision, revealing how photosynthetic complexes are organised and settling long-standing debates on chloroplast architecture.
buff.ly/j3TSIkn
20.09.2025 13:59 — 👍 68 🔁 18 💬 3 📌 7
A new #UExM probe for #Actin!
Fun times thinking/developing/testing with @centriolelab.bsky.social & @lreymond.bsky.social !
Congrats to all !
If interested in #UExM, or #HAK-Actin, feel free to reach out !
Soon available at @spirochrome.com
#Expansion #Microscopy #ProtistsOnSky
28.08.2025 07:16 — 👍 45 🔁 19 💬 3 📌 0
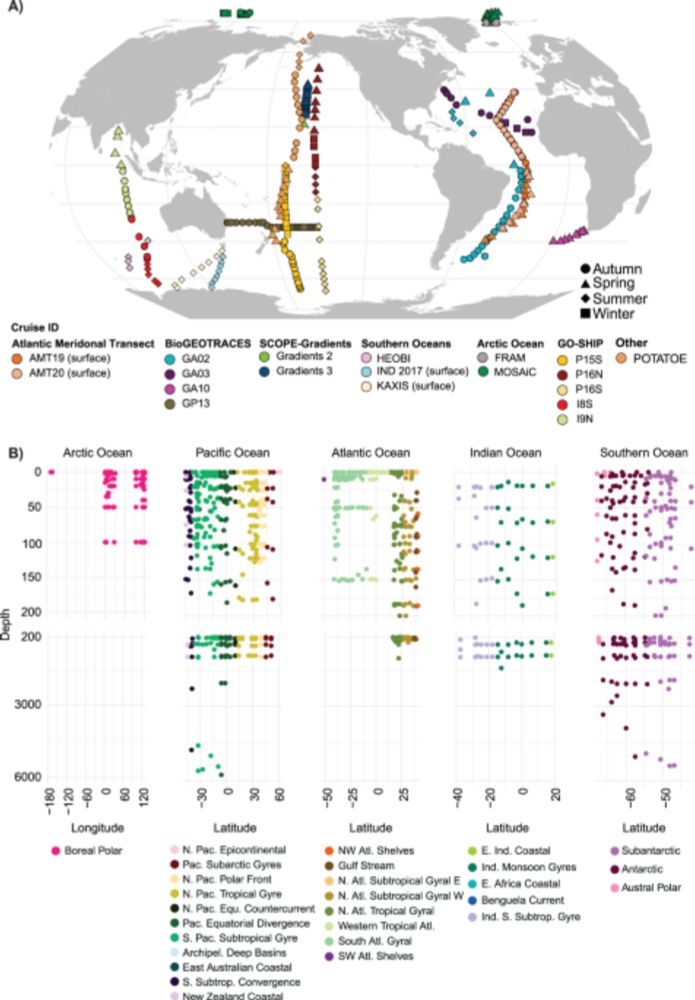
Characterizing organisms from three domains of life with universal primers from throughout the global ocean - Scientific Data
Scientific Data - Characterizing organisms from three domains of life with universal primers from throughout the global ocean
Please Re-Post. Our global GRUMP microbial ocean database is out in Scientific Data! Unfractionated, single universal PCR, with Archaea, Bacteria, and Eukaryotes all on the same scale with the same denominator. Pole to pole, depths to 6000m. Lots of metadata. www.nature.com/articles/s41...
02.07.2025 16:19 — 👍 45 🔁 31 💬 2 📌 2

After 5 amazing years @pilhoferlab.bsky.social I'm starting my lab @embl.org Grenoble this November embl.org/wollweber
We'll image Asgard archaea and many other strange microbes ( #archaeasky, #protistsonsky..) to understand eukaryogenesis
First job ad: #teamtomo scientist!🔬❄️ tinyurl.com/2rdu2ze6
27.06.2025 10:58 — 👍 159 🔁 45 💬 9 📌 5
Excited to share the lab's 1rst preprint! Rapid high-res immunofluorescence is now possible in cultured & environmental diatoms thanks to 4-fold expansion microscopy. A step-change for comparative cell biology in one of the most important phytoplankton groups on the planet 🥳
tinyurl.com/m9s5su7s 1/2
17.06.2025 07:33 — 👍 323 🔁 89 💬 8 📌 19
Final PhD paper now reviewed and published in JSB:X. I was happy with some great reviews that improved the paper! Thanks to Sander Roet for his help with the code base, and Remco Veltkamp and @fridof.bsky.social
09.06.2025 06:56 — 👍 20 🔁 4 💬 0 📌 1
Are you a recent dinoflagellate convert, or already long dedicated to the craft? Don't you wish you could talk about them more?
Join Manon's DinoSphere seminar series! All things phylogeny, imaging, ultrastructure and more :)
#protistsonsky
04.06.2025 05:52 — 👍 6 🔁 0 💬 0 📌 0
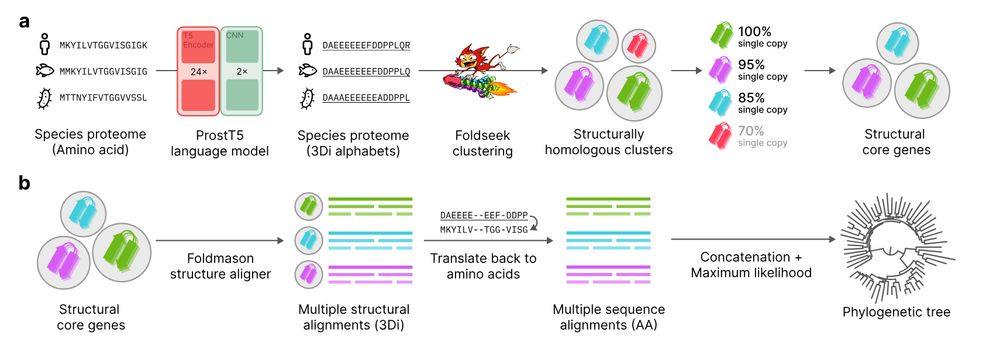
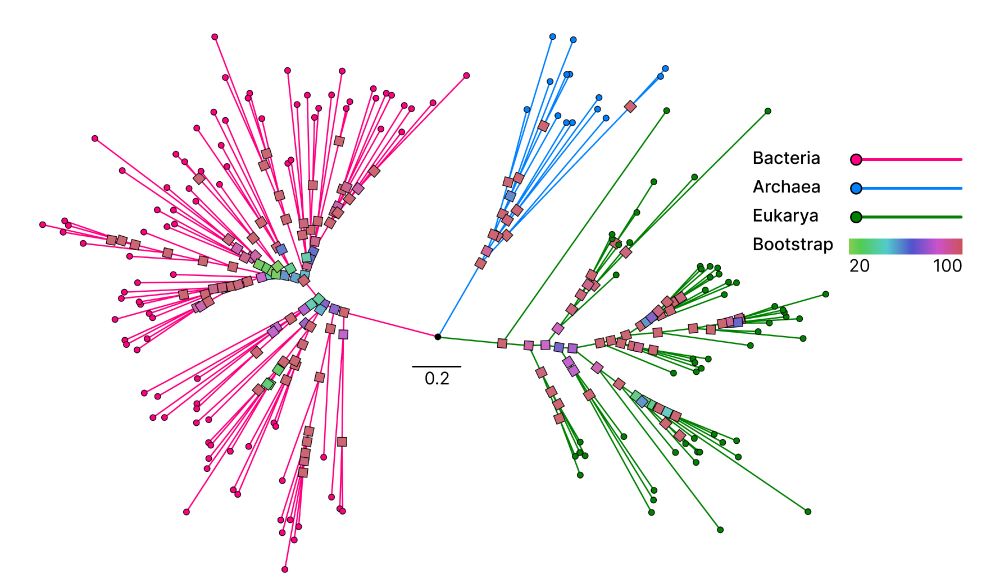
Unicore identifies single-copy protein structures across genomes using Foldseek, bypassing slow structure predictions by utilizing 3Di predictions from ProstT5, enabling rapid phylogenetic inference at the tree-of-life scale. 1/n
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
23.12.2024 16:39 — 👍 121 🔁 57 💬 2 📌 3
Anti-gatekeeping workflows is what keeps #teamtomo growing! Love to see it.
07.05.2025 15:16 — 👍 13 🔁 1 💬 0 📌 0
Lucky to be a PDRA in @mackinderlab.bsky.social. Interested in all things pyrenoid and some things phase separated.
https://james-r-barrett.github.io/
Geomicrobiologist 🌋 at @embl.org studying extreme microbial ecosystems in the Central Andes
Biological oceanographer & phytoplankton connoisseur. Forest Lover. Stand up #TEDx person.🧚🏼♀️🦄🔬🌊 Views=mine. @institutrb.bsky.social
@BigelowLab.bsky.social @EMBO_YIN
https://jelenagodrijan.wixsite.com/godi
Investigating mechanisms of eukaryotic cellular quality control with a focus on autophagy @MPIbp via #cryoET, #teamtomo, #cellbiology and #massspectrometry
I like to think about how cells work, especially among #protists. Driven by curiosity and a love to share detailed research and odd observations. Senior Research Scientist, Bigelow Laboratory for Ocean Sciences. He/him. https://www.protistsystems.org/
Neuroscientist and Super-Resolution Microscopist
Biomedical animation at WEHI, Melbourne Australia
Animation playlist
https://www.youtube.com/playlist?list=PLD0444BD542B4D7D9
Molecular Cell is a Cell Press journal that aims to publish the best papers in molecular biology.
Prof of Computational Microbiology. Oxford Biochemistry & St Anne’s College. Bacterial Cell Envelopes. LFC fan 🌈
The Young Scientific Symposium is a student-led scientific conference focused on the interface of chemistry and biology. Organized by PhD students and postdocs, the symposium brings together students and early-career researchers.
IECB - BORDEAUX
Connecting the Latin American bioimaging community by establishing a robust collaborative network that facilitates access to training and research infrastructure.
Boldly exploring our unknown Ocean
Professor of microbial ecology and evolution at Univ of Texas Austin.
Postdoc in the Baum lab at the MRC-LMB, studying the diversity and behaviour of Asgard archaea
Post-doc at the @CentrioleLab in Geneva
Our main focus is to offer innovative tools for live cell fluorescence imaging to the scientific community.
https://spirochrome.com/
Professor at Stockholm University, Sweden. Weekend violinist. We study how enzymes work and how biological systems capture energy.
CryoEM, membrane proteins and whatnot

















