Announcement for the 6th Protein Engineering Canada Conference, to be held June 22nd-24th in Ottawa, Canada. The image shows a protein structure in front of a picture of Parliament Hill and Chateau Laurier in Ottawa.
1/
Save the date!
The 6th Protein Engineering Canada Conference will be held on June 22-24 in Ottawa, Canada.
Abstract submission and registration are open!
More information here: event.fourwaves.com/pec2026/pages
04.12.2025 16:32 — 👍 4 🔁 3 💬 1 📌 0

2/
Speakers of the 6th Protein Engineering Canada Conference include:
Bill DeGrado
Joelle Pelletier
Tim Whitehead
Anastassia Vorobieva
Lucy Colwell
Ai Niitsu
@joannas.bsky.social
@nickpolizzi.bsky.social
@possuhuanglab.bsky.social
@paolalaurino.bsky.social
@stephanhammer.bsky.social
and more!
04.12.2025 16:32 — 👍 3 🔁 2 💬 0 📌 0

Announcement for the 6th Protein Engineering Canada Conference, to be held June 22nd-24th in Ottawa, Canada. The image shows a protein structure in front of a picture of Parliament Hill and Chateau Laurier in Ottawa.
1/
Save the date!
The 6th Protein Engineering Canada Conference will be held on June 22-24 in Ottawa, Canada.
Abstract submission and registration are open!
More information here: event.fourwaves.com/pec2026/pages
04.12.2025 16:32 — 👍 4 🔁 3 💬 1 📌 0

Enzyme-like proteins by computational design | PNAS
We report the development and initial experimental validation of a
computational design procedure aimed at generating enzyme-like protein
catalys...
Similarly, enzyme function can be designed de novo by creating a new active site within a natural protein scaffold that lacks the target activity, even if that catalytic function exists in nature.
See below for an early example:
www.pnas.org/doi/full/10....
31.07.2025 22:10 — 👍 2 🔁 0 💬 0 📌 0
Overall, our study:
✅ Introduces a new strategy to transform minimal protein scaffolds into biocatalysts
✅ Provides mechanistic insights from crystallography & molecular dynamics
✅ Opens the door to designing custom lids for more complex reactions, which we’re now exploring
Thanks for reading! 🧵🧬
29.07.2025 18:33 — 👍 2 🔁 0 💬 1 📌 0
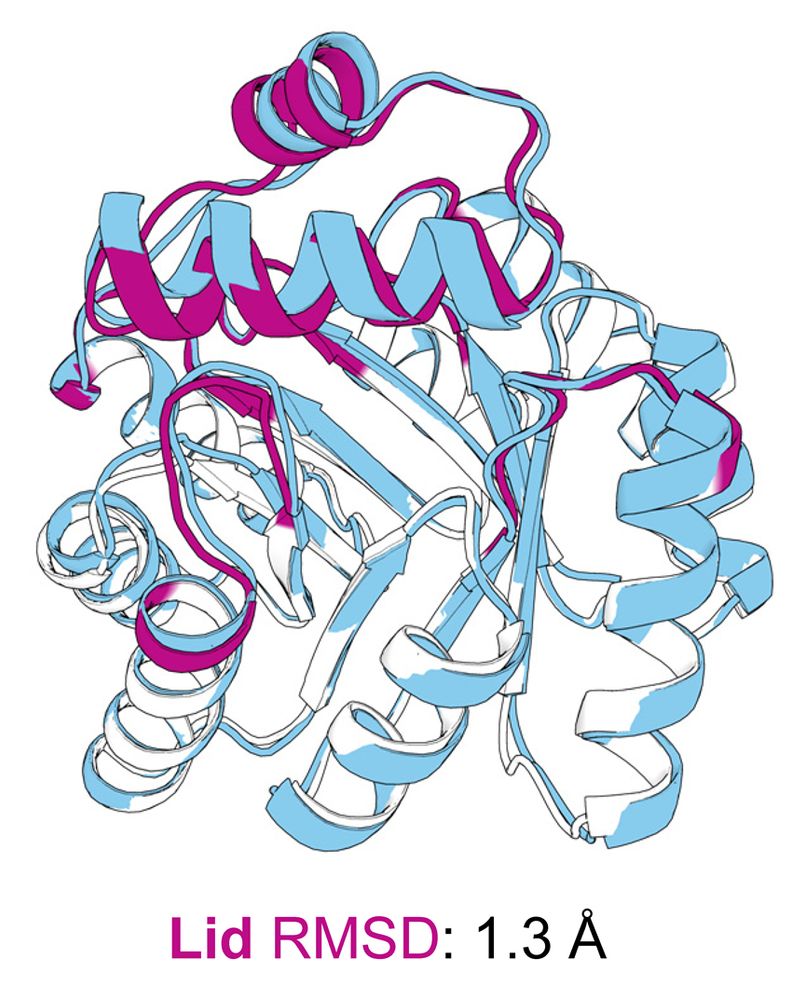
The crystal structure (blue) aligns closely with the design model (minimal TIM barrel and lid colored white and magenta, respectively).
Our crystal structure validated the designed fold, confirming that the lid was correctly folded.
However, a subtle 1.8 Å lid shift disrupted a key catalytic contact, likely contributing to the modest activity. But structural analysis reveals paths to improve activity in the next round of design!
29.07.2025 18:33 — 👍 2 🔁 0 💬 1 📌 0
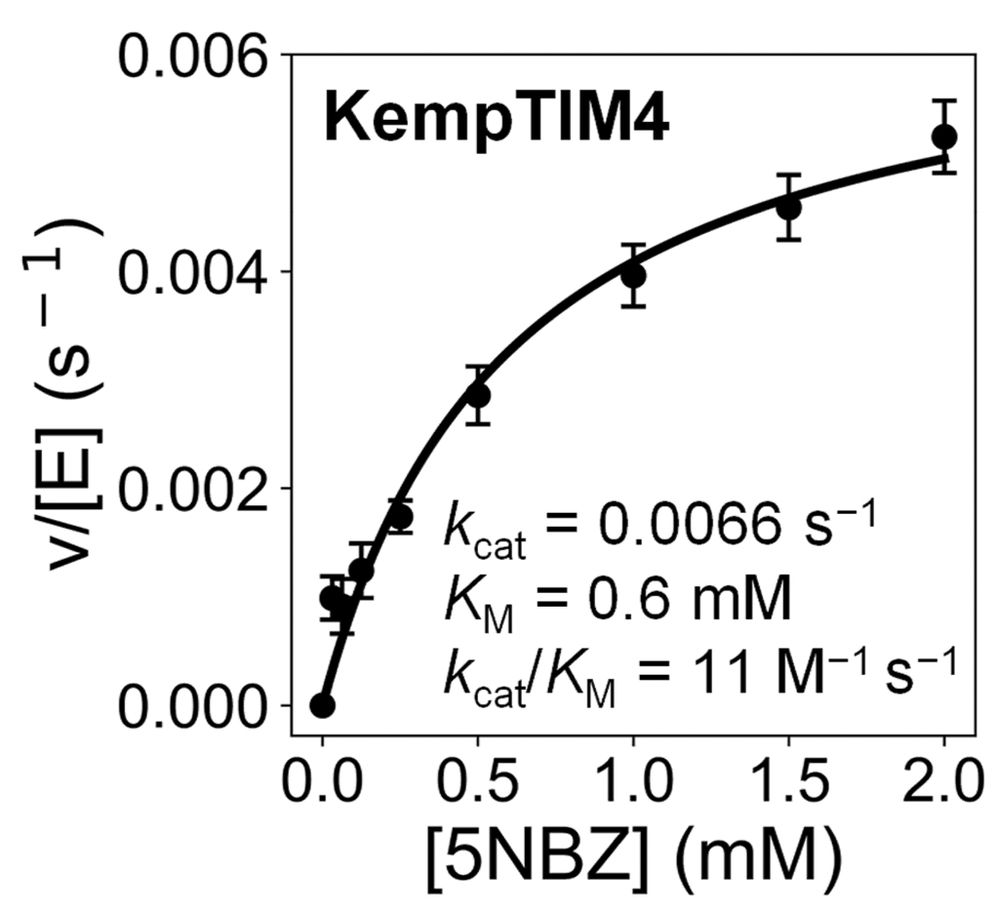
Michaelis-Menten plot of KempTIM4 showing saturation kinetics.
One of our designs, KempTIM4, showed catalytic efficiency comparable to many first-round de novo Kemp eliminases generated by traditional methods.
29.07.2025 18:33 — 👍 1 🔁 0 💬 1 📌 0
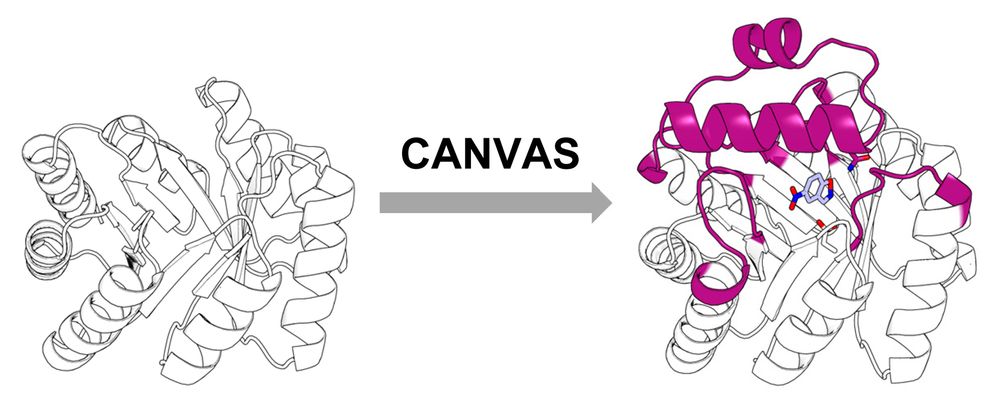
Building a custom lid onto a minimal, de novo TIM barrel using CANVAS.
Using CANVAS, we designed a structural lid onto a minimal, de novo TIM barrel to anchor catalytic residues and create an active site for the Kemp elimination reaction.
29.07.2025 18:33 — 👍 1 🔁 0 💬 1 📌 0
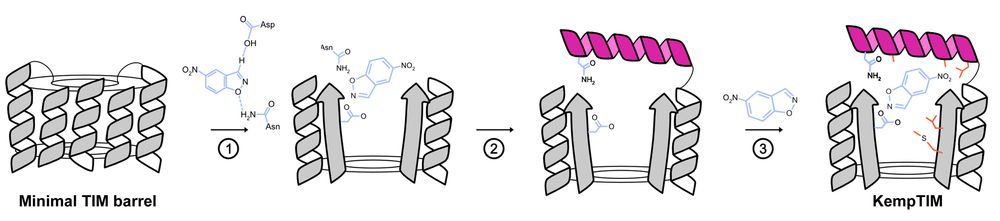
De novo enzyme design using CANVAS.
TIM barrels are among nature’s most powerful enzyme scaffolds but making them from scratch with catalytic function has been a challenge.
Enter CANVAS: a computational pipeline combining Triad, RFdiffusion & ProteinMPNN to customize minimal TIM barrels into functional enzymes.
29.07.2025 18:33 — 👍 1 🔁 0 💬 1 📌 0
Congratulations! Looking forward to seeing all the exciting science that will come out of your lab! 🧪
25.07.2025 15:50 — 👍 1 🔁 0 💬 0 📌 0

Research Position (PhD) in organic chemistry and b...
<div style="text-align: justify;">The research group „Organic Chemistry and&nb...
Join us! We are looking for a new team member (PhD student) with strong background in organic chemistry.
🙏 RETWEET (We want to recruit internationally)
Organic chemistry meets #DirectedEvolution
Highly interdisciplinary & passionate research group
uni-bielefeld.hr4you.org/job/view/433...
27.06.2025 21:13 — 👍 6 🔁 7 💬 0 📌 0
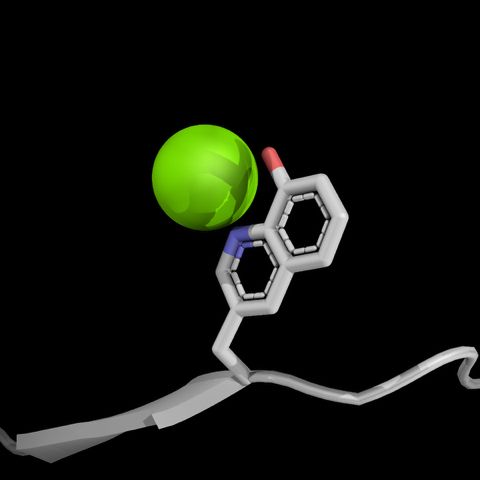
Non-canonical amino acid from PDB ID 8W3Z shown chelating to a magnesium ion. Image made with PyMol.
https://www.rcsb.org/structure/8W3Z
https://www.pymol.org
Protein Engineering, Design & Selection (PEDS) invites contributions to a Special Collection titled, “Non-Canonical Amino Acids", with guest editors Prof. Huiwang Ai (Virginia) and Prof. Peng Chen (Peking). Send us your best work!
academic.oup.com/peds/pages/c...
15.05.2025 00:25 — 👍 12 🔁 4 💬 0 📌 0

Guess what? TPS has extended the deadline to March 19 to submit abstracts for poster presentations and speaking opportunities at our 39th Annual Symposium. Join us in San Francisco June 26 - 29 for 3.5 days of scientific talks.
hashtag#proteinscience hashtag#annualsymposium
lnkd.in/g7VKqX7C
06.03.2025 18:27 — 👍 3 🔁 5 💬 0 📌 0
Molecular dynamics simulations showed that distal mutations enhance active-site accessibility—either by loosening loops covering the active site or widening bottlenecks for substrate entry & product exit. The enzyme breathes more efficiently! 🌬️ (5/6)
28.02.2025 17:17 — 👍 0 🔁 0 💬 1 📌 0

Kinetic solvent viscosity effects & stopped-flow experiments showed that distal mutations don’t just tweak structure—they accelerate substrate binding & product release. (4/6)
28.02.2025 17:17 — 👍 0 🔁 0 💬 1 📌 0
Crystal structures showed that active-site mutations pre-organize the catalytic machinery. But distal mutations? They subtly tune conformational dynamics—enhancing productive substates & reshaping the energy landscape of the catalytic cycle. (3/6)
28.02.2025 17:17 — 👍 0 🔁 0 💬 1 📌 0
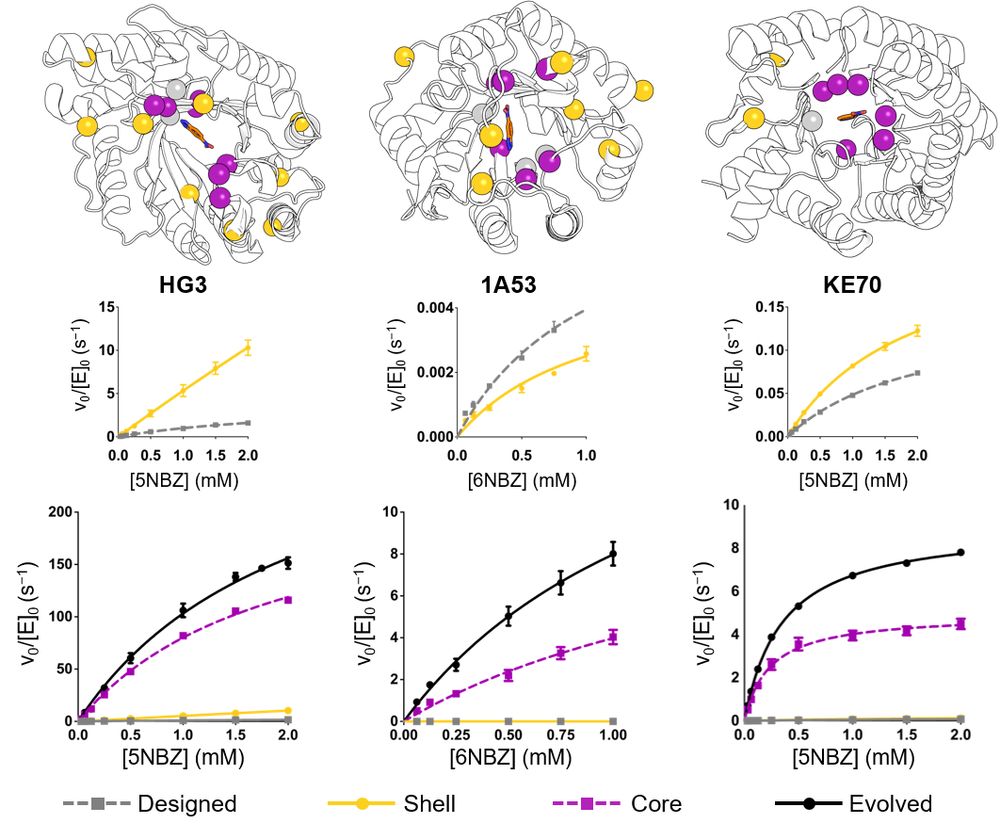
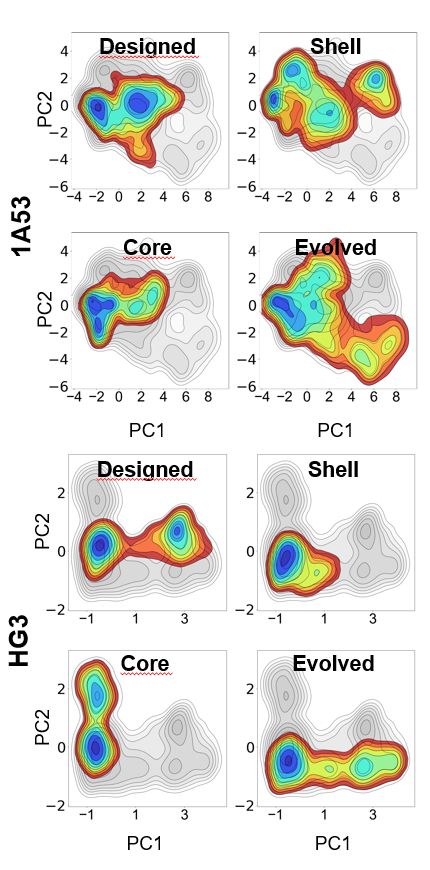
We engineered "Core" and "Shell" variants of three evolved Kemp eliminases to dissect the effects of active-site vs. distal mutations. Core mutations dramatically boosted catalysis. Shell mutations alone? Not much—until they worked together in evolved enzymes. 🔍 (2/6)
28.02.2025 17:17 — 👍 1 🔁 0 💬 1 📌 0
How do mutations far from an enzyme's active site influence catalysis? 🤔
Part 2: In collaboration with @fraserlab.bsky.social and @silviaosuna.bsky.social, we investigated this question using de novo Kemp eliminases, revealing effects of distal mutations on the catalytic cycle. 🧵 (1/6)
28.02.2025 17:09 — 👍 6 🔁 2 💬 1 📌 0
Thank you for the feedback!
28.02.2025 16:13 — 👍 1 🔁 0 💬 0 📌 0
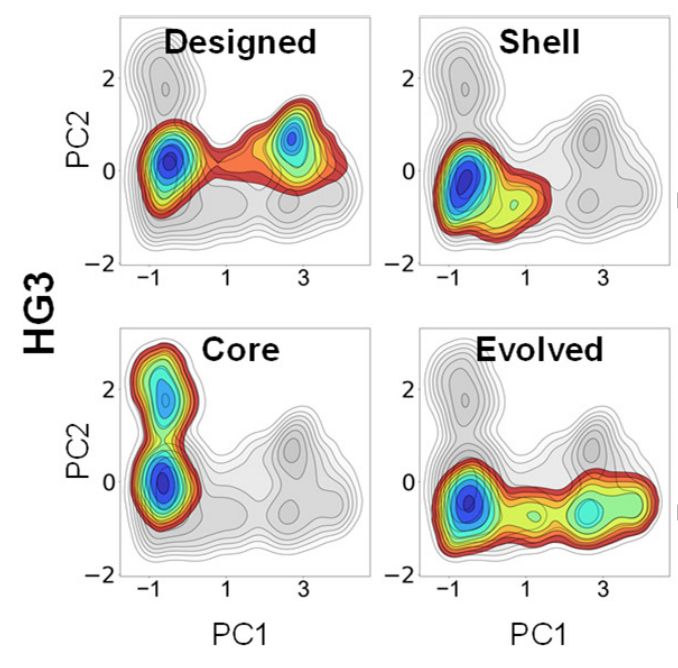
Figure from https://www.biorxiv.org/content/10.1101/2025.02.21.639315v1.full.pdf showing how mutations to the active site (labeled "core" relative to the parental, which is labeled "designed"), to second-shell residues (labeled "shell"), or both (labeled "evolved") affect the conformational dynamics of a de novo designed kemp eliminase.
Whereas beneficial active site mutations to enzymes often improve the chemical transformation itself by preorganizing the active site, mutations to second-shell residues instead tune steps like product release by modifying the broader conformational ensemble www.biorxiv.org/content/10.1...
28.02.2025 09:35 — 👍 8 🔁 1 💬 1 📌 0
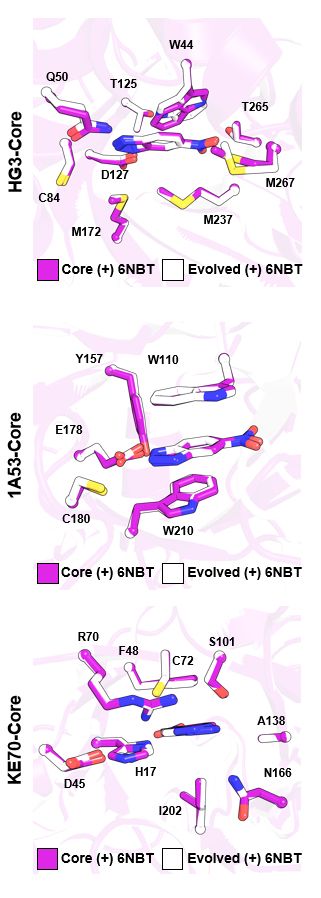
Mechanistically, distal mutations:
🔹 Alter loop flexibility
🔹 Reshape enzyme motions to favor product release
🔹 Realign local electric fields to lower the barrier for C-C bond cleavage
These changes shift the rate-limiting step from C-C bond cleavage to product release—and speed it up!
27.02.2025 21:16 — 👍 2 🔁 0 💬 1 📌 0
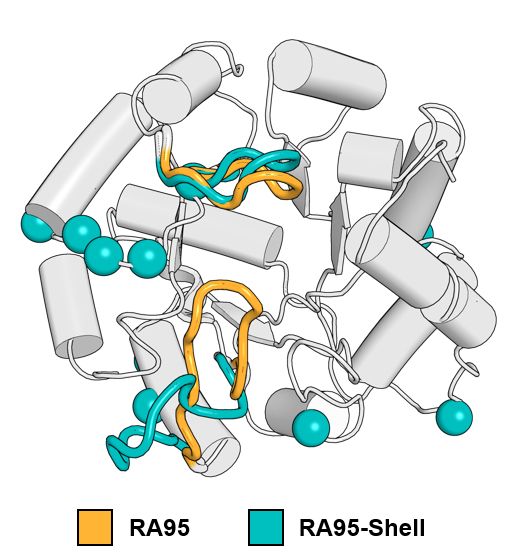
Crystal structures revealed that distal mutations trigger large-scale conformational shifts in an active-site loop, making the active site more open. Interestingly, these mutations aren't located on the loop—making their effects hard to predict!
27.02.2025 21:16 — 👍 1 🔁 0 💬 1 📌 0
Origins of life, astrobiology and synthetic biology group at Charles University in Prague
An Inorganic Design, Computational Chemistry, Machine Learning, and Enzyme Catalysis Research Group in Chemical Engineering at MIT.
Led by Prof. Heather J. Kulik
hjkgrp.mit.edu
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique
Computational chemistry and AI for drug design in biotech #compchem | CSIRO, Melbourne AU
MIT biology lab using computational and experimental methods to study protein structure, function, and interactions
PhD student @MIT • Research on Generative Models for Biophysics and Drug Discovery
Chair of Sustainable Biomaterials (glycoscience, ice and polymers) in Department of Chemistry and and Institute of Biotechnology, University of Manchester. Co-founder @Cryologyx
News from the King Lab kinglab.ipd.uw.edu.
Part of @uwmedicine.bsky.social and @uwproteindesign.bsky.social.
Some assembly required.
Schmidt Science Fellow | Postdoc @ Stanford | Prev. DPhil @ Oxford || AI for Molecule & Cell Modeling
Computational designer of molecular tools for intracellular signaling. Assistant Professor at UCLA Bioengineering. Website: www.jasonzhanglab.com Lab’s Bluesky: https://bsky.app/profile/jasonzhanglab.bsky.social
Avid rower who sometimes thinks about biomolecular dynamics. Asst Prof @uwbiochem.bsky.social
https://waymentsteelelab.org/
Protein Engineering and Design- Synthetic Biology- Science Education
Postdoc @ EPFL. Previously @ ENS and Institut Pasteur. Protein design, Protein Language Models.
Protein Computational Biology Lab @ UFMG
We use and develop computational and biophysical methods to study protein structure, function and evolution.
P.I.: Lucas Bleicher
Lab head at the University of Melbourne. Studying the structure and biochemistry of microbial membrane proteins and metalloenzymes.
Professor detached@CNRS/Sorbonne Université, director of CQSB lab UMR7238, ERC and IUF fellow, co-founder@qubit_pharma, structural bioinformatics, Chemoinformatics, molecular visualization