Assistant Professor (Tenure Track) of Computational Immunology
My department @bsse.ethz.ch is inviting applications for a new assistant professor (tenure-track) in computational immunology, interested candidates please see advert and apply! Deadline 15 April 2026 ethz.ch/en/the-eth-z...
16.02.2026 08:35 — 👍 0 🔁 2 💬 0 📌 0
New preprint alert: we use sign errors as a test of how well TWAS works.
Very worryingly we find that TWAS gets the sign wrong around 1/3 of the time (compared to 50% for pure guessing). You can read more about our analysis here, and what we think is going on 👇
06.01.2026 02:48 — 👍 67 🔁 28 💬 5 📌 0
Importantly in this paper we derive a new metric PRS pleiotropy which we use to show shallow phenotypes indeed give non specific gwas signal that leads to non specific PRS predictions
20.12.2025 15:44 — 👍 1 🔁 0 💬 2 📌 0

Embryo selection company Herasight goes all in on eugenics
...
I wrote about the bizarre case of Herasight, the embryo selection company going all in on eugenics.
13.12.2025 20:15 — 👍 126 🔁 84 💬 6 📌 15

The PGC Suicide Working Group will provide a symposium at #WCPG2025 🇲🇽 covering our latest multi-ancestry GWAS and CNV meta-analyses, sex-specific meta-analyses, and GxEHR analyses. 🥳
@lcstoshio.bsky.social @andreyshabalin.bsky.social @sarahcolbert.bsky.social @caina89.bsky.social
19.10.2025 18:17 — 👍 17 🔁 5 💬 2 📌 0

We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
22.09.2025 05:29 — 👍 175 🔁 90 💬 4 📌 5

🌎👩🔬 For 15+ years biology has accumulated petabytes (million gigabytes) of🧬DNA sequencing data🧬 from the far reaches of our planet.🦠🍄🌵
Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
03.09.2025 08:39 — 👍 218 🔁 118 💬 3 📌 16

Postdoctoral Scholar – Genomics, Aging, Somatic Mutation, Structural Variation, Evolution , Cancer – Integrative Biology
University of California, Berkeley is hiring. Apply now!
The Sudmant lab at UC Berkeley is seeking a postdoc to work on a fully funded NIH project to understand differences in DNA repair and somatic mutation across the primate tree of life. Please spread widely to those who may be interested aprecruit.berkeley.edu/JPF05052
13.08.2025 04:22 — 👍 55 🔁 55 💬 0 📌 1
Home - ProbGen 2026
Your Site Description
The 2026 Probabilistic Modeling in Genomics (ProbGen) meeting will be held at UC Berkeley, March 25-28, 2026. We have an amazing list of keynote speakers and session chairs:
probgen2026.github.io
Please help spread the news.
06.06.2025 17:52 — 👍 69 🔁 36 💬 2 📌 0
Congratulations, very exciting! 🎉
26.07.2025 05:54 — 👍 1 🔁 0 💬 0 📌 0
‘Genetic Risk Effects on Psychiatric Disorders Act in Sets’, the title of our new preprint on
@medrxivpreprint.bsky.social! This huge collaborative effort advances our understanding of psychiatric genetic architecture and emphasizes the importance of looking beyond additive effects. 🧵1/n; 🧪👩🏽🔬🧬
24.07.2025 08:15 — 👍 10 🔁 8 💬 1 📌 1
This work would not have been possible if not for the persistent efforts of @jolienrietkerk.bsky.social Andy Dahl, Jonathan Flint, Andrew Schork and many others, as well as the data from participants in @ukbiobank.bsky.social and IPSYCH. We hope it will be informative to the field. 12/n
24.07.2025 05:24 — 👍 2 🔁 0 💬 1 📌 0
Though our investigations and findings are centered on psychiatric disorders, the implications are generalizable to all complex traits and diseases, especially those with heterogeneous architectures and unclear diagnostic boundaries. 11/n
24.07.2025 05:24 — 👍 3 🔁 0 💬 1 📌 0
Overall, our work provides a novel metric, the CE test, for informing diagnostic boundaries. Our results show that genetic effects on psych disorders act in sets, and calls for a re-evaluation of current approaches and assumptions. 10/n
24.07.2025 05:24 — 👍 3 🔁 0 💬 1 📌 0
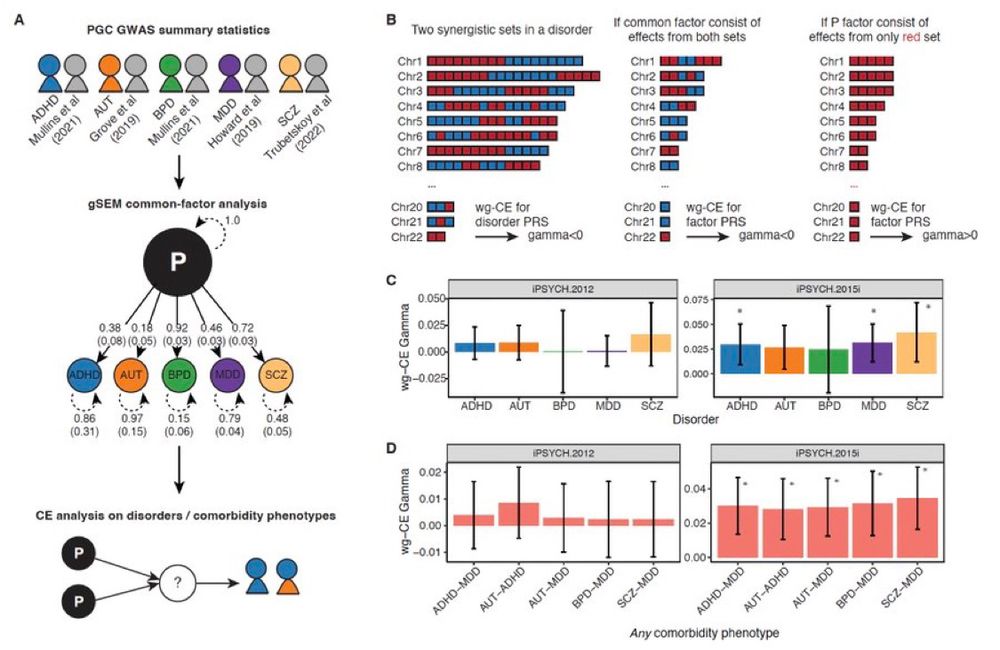
Finally, we find that common genetic effects across all five psych disorders, expected to capture common etiological axes among them, forms a cross-order set most plausibly explained by common confounders external to each disorder’s etiology. 9/n
24.07.2025 05:24 — 👍 8 🔁 0 💬 4 📌 1
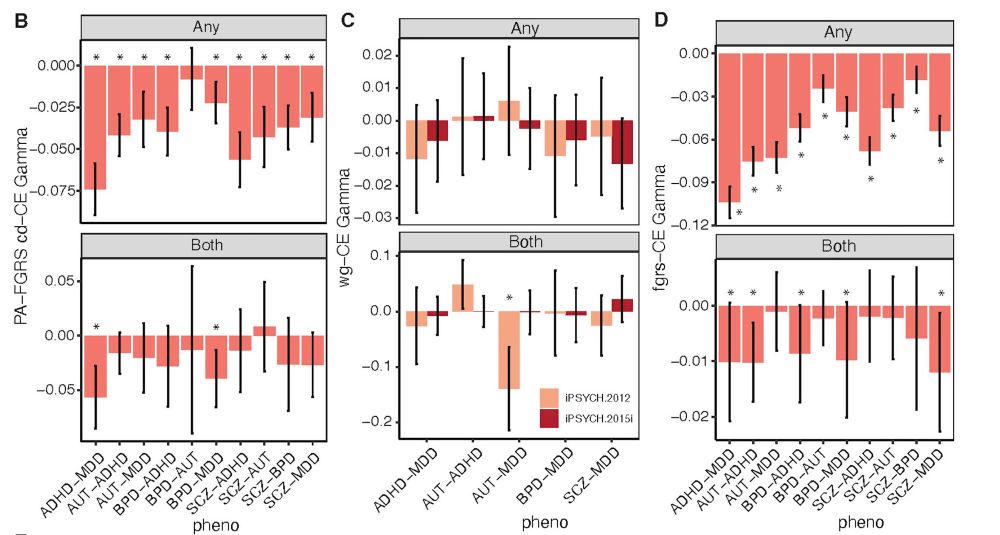
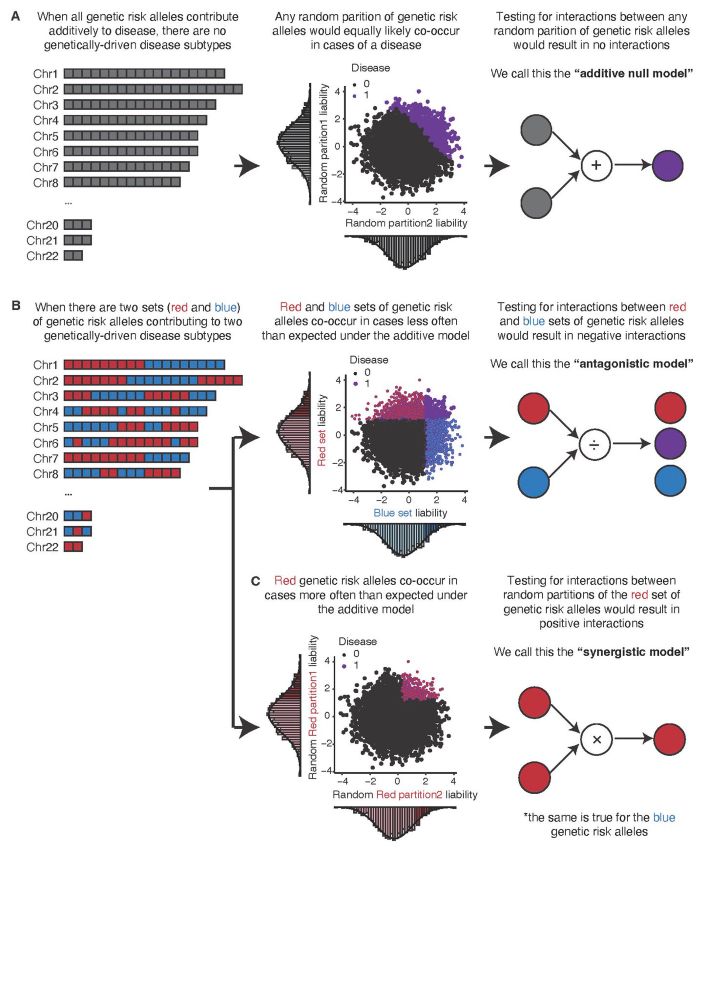
We further show that disorder-specific sets lead to comorbidity between two disorders, refuting the assumption that comorbidity is the result of additive effects of genetic effects on both, exemplified by the expectation that high rGs would lead to high comorbidity. 8/n
24.07.2025 05:24 — 👍 3 🔁 0 💬 1 📌 0
These findings imply that rGs are insufficient to inform nosology, and that the CE test provides a way to determine whether putative disorders or subtypes should be merged or remain separate diagnostically. 7/n
24.07.2025 05:24 — 👍 5 🔁 0 💬 1 📌 0
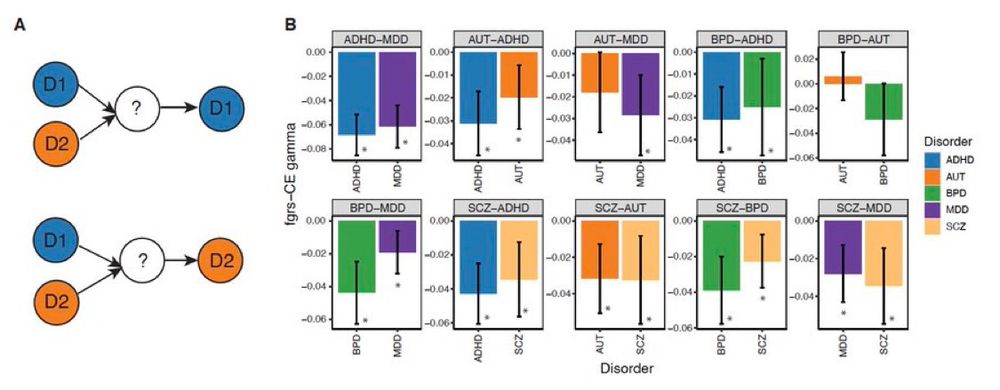
We also show that sets of genetic effects are disorder-specific, despite high genetic correlations (rG) between them. This opposes a well-established conjecture, based on high rG, that clinical boundaries do not reflect their pathogenic processes. 6/n
24.07.2025 05:24 — 👍 5 🔁 1 💬 1 📌 0
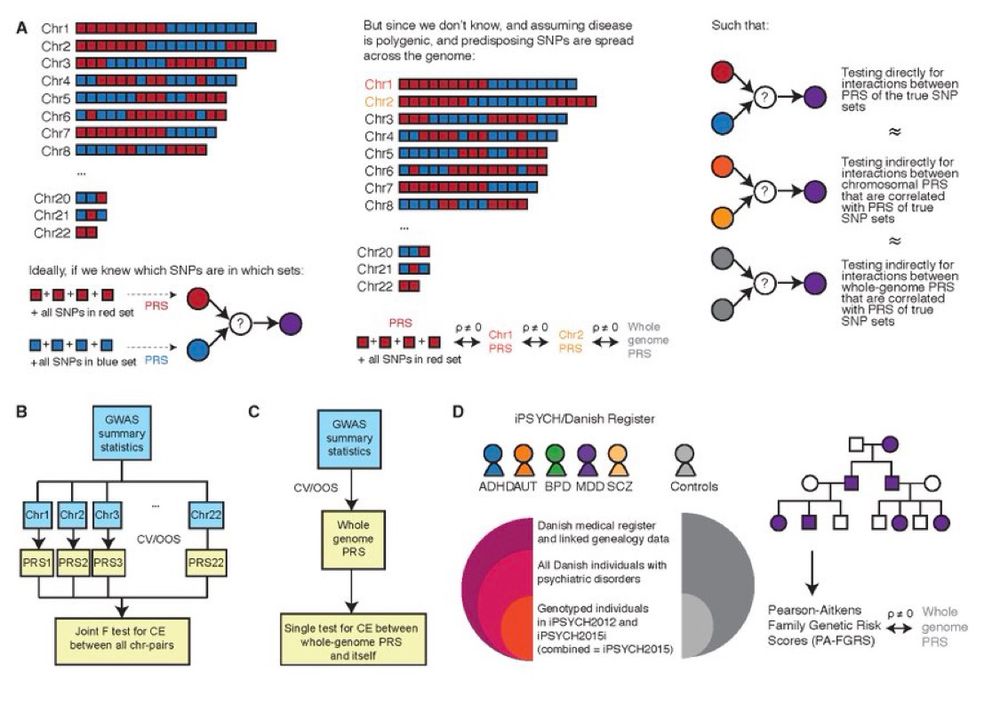
In our paper, we test for CE in five psychiatric disorders in the @ukbiobank.bsky.social and the iPSYCH dataset using both polygenic risk scores (PRS) and family-based genetic risk scores (FGRS). We find negative CE for 3 out of 5 disorders, demonstrating there are unrecognized subtypes. 5/n
24.07.2025 05:24 — 👍 2 🔁 0 💬 1 📌 0

A model and test for coordinated polygenic epistasis in complex traits | PNAS
Interactions between genetic variants—epistasis—is pervasive in model systems and
can profoundly impact evolutionary adaption, population disease d...
The existence of these sets induces a structured form of statistical interactions called coordinated epistasis (CE), detailed in previous publications by Andy Dahl, Noah Zaitlen www.pnas.org/doi/10.1073/...; negative CE is seen when there are different sets of genetic effects. 4/n
24.07.2025 05:24 — 👍 4 🔁 0 💬 1 📌 0
These sets, should they exist, would each drive an unrecognized etiological subtype. Our paper tests for the existence of these sets in five psychiatric disorders. 3/n
24.07.2025 05:24 — 👍 2 🔁 0 💬 1 📌 0
Our paper addresses a central question in quantgen: whether all genetic variants associated with a complex disease contribute additively to its liability as commonly assumed, or there exists sets of them that co-occur more in certain cases than expected under additivity. 2/n
24.07.2025 05:24 — 👍 4 🔁 0 💬 1 📌 0
Very happy to share our new paper now on @medrxivpreprint: “Genetic risk effects on psychiatric disorders act in sets”, a great effort led my PhD student @jolienrietkerk.bsky.social, and performed together with collaborators Andy Dahl, Jonathan Flint, Andrew Schork etc. Thread 1/n
24.07.2025 05:24 — 👍 44 🔁 10 💬 3 📌 0
PhD student at Bioinformatics and Cellular Genomics group @SVIResearch
Computational and statistical genetics. Excited about all things science. Professor of genetics at UPenn. Views my own.
https://www.med.upenn.edu/bogdan-group/
Statistical Geneticist, Group leader at University of Lausanne/Unisante, father, climber, runner
assistant professor at ucsf interested in genetics, statistics, etc…
jeffspence.github.io
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
Disease genomics & molecular mechanisms; ME/cfs: http://decodeme.org.uk, SequenceME @ Edinburgh University. Views - my own. He/him.
Assistant Professor, Cold Spring Harbor Laboratory
Systems immunology to understand thymus physiology and T cell development
@RingAScientist, @SkypeScientist
Human Genetics, Molecular Epidemiology, Biological Psychiatry
https://medicine.yale.edu/lab/polimanti/
Head of Estonian Biobank & Professor of Pharmacogenomics at the Estonian Genome Centre, University of Tartu.
AI slop (except one) enjoyer
Dad. Associate Professor @ ETH Zurich. We study stress, behavior, hippocampus, noradrenaline and the mighty locus coeruleus. We work with mice and focus a lot on 3Rs.
Here to learn, share, laugh and rant.
Evolutionary and statistical geneticist. Associate Professor at
Center for Genomic Sciences, UNAM.
http://sohaillab.com
UNAM | U Chicago | Langebio | Harvard | MIT
🇵🇰🇺🇸🇲🇽🏳️🌈
Associate Professor @ UC-Davis | Former Senior Director @ Exai Bio | Computational Biology, ML/AI, Genomics, RNA, Cancer
Director of the Institute of AI for Health @ Helmholtz Munich
Assistant Professor at @EPFL in Computer Science and Life Sciences. PostDoc at @Genentech and @Stanford. PhD at @ETH and @BroadInstitute.
www.aimm.epfl.ch
computational genomics prof at med uni innsbruck. co-created the michigan imputation server. co-created two kids ❤️.
interested in AI, genomics, chronic diseases, nextflow, complex gene regions
https://genepi.i-med.ac.at/team/schoenherr-sebastian
MD. PhD. Assistant Professor @ETH Zurich. Previously @Stanford CS.
This is the official account of the ETH-Department of Biosystems Science and Engineering (D-BSSE). https://bsse.ethz.ch