Not yet, but we will surely generate bp-resolution genome-wide scores for all six species studied in the paper and make them publicly available. For now, we have predictions for ~10M variants used in the S-LDSC analysis in humans.
22.09.2025 14:59 — 👍 3 🔁 0 💬 0 📌 0
This is truly an incredible breakthrough IMO. Really exemplifies what you get when deep domain expertise (popgen/evolution/disease genetics in this case) fuses with cleverly crafted ML. What u get r sleek, well thought out architectures that absolutely destroy the behemoths. Wow!! 1/
22.09.2025 08:34 — 👍 59 🔁 14 💬 1 📌 1
All in all, we believe that GPN-Star offers a scalable & flexible approach for training effective gLMs.
This work was led by my talented students @czye.bsky.social and @gonzalobenegas.bsky.social, with contributions from other lab members, @peterdfields.bsky.social at Jax, & B. Clarke at DKFZ
(n/n)
22.09.2025 05:29 — 👍 4 🔁 1 💬 1 📌 0

GitHub - songlab-cal/gpn: Genomic Pre-trained Network
Genomic Pre-trained Network. Contribute to songlab-cal/gpn development by creating an account on GitHub.
Upon publication, we will release base-resolution predictions for the human genome and the five model organisms.
Codes to train the model, run inference, and reproduce the analyses are available on GitHub (github.com/songlab-cal/...) and Hugging Face (tinyurl.com/nhhcppvm).
(9/n)
22.09.2025 05:29 — 👍 7 🔁 0 💬 1 📌 0

To show that GPN-Star is a robust and generalizable framework that can advance biology beyond human genetics, we apply it to train gLMs for five well-studied model organisms and demonstrate their effectiveness in assessing variant effects in these species.
(8/n)
22.09.2025 05:29 — 👍 4 🔁 0 💬 1 📌 0

In addition, GPN-Star exhibits meaningful nucleotide dependencies that align with known functional dependencies, indicating its potential to help understand genomic syntax. This represents a notable advance over traditional conservation scores.
(7/n)
22.09.2025 05:29 — 👍 7 🔁 0 💬 1 📌 0


By training GPN-Star on vertebrate, mammal, and primate alignments, we reveal task-dependent advantages of modeling deeper versus more recent evolution. These findings offer new biological insights and practical guidance for developing future gLMs and evolutionary models.
(6/n)
22.09.2025 05:29 — 👍 4 🔁 2 💬 1 📌 0

GPN-Star achieves unprecedented SNP heritability enrichments across over 100 human complex traits. Moreover, we devise a simple approach to incorporate tissue-specificity into the model prediction and show that it further improves heritability enrichment.
(5/n)
22.09.2025 05:29 — 👍 4 🔁 0 💬 1 📌 0

We compare GPN-Star with several models, including the recent AlphaGenome and Evo2 models with up to 1Mb context size and 40B parameters, and observe that GPN-Star consistently ranks at the top across a wide range of human variant effect prediction tasks.
(4/n)
22.09.2025 05:29 — 👍 3 🔁 0 💬 1 📌 0
We also introduce a calibration method that removes the confounding effect of mutation rate variation from gLM predictions for the first time. This improves downstream performance and enables a more direct interpretation of model scores as estimates of selective constraint.
(3/n)
22.09.2025 05:29 — 👍 5 🔁 1 💬 1 📌 0

GPN-Star features a novel phylogeny-aware architecture that enables the model to explicitly capture evolutionary relationships encoded in whole-genome alignments and overcomes the key limitations of our earlier model GPN-MSA (doi.org/10.1038/s415...).
(2/n)
22.09.2025 05:29 — 👍 8 🔁 0 💬 1 📌 2

We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
22.09.2025 05:29 — 👍 169 🔁 89 💬 4 📌 5
Thanks, Josh. I wish you had been one of our reviewers—life would’ve been so much easier.
11.09.2025 04:45 — 👍 2 🔁 0 💬 0 📌 0
Hi Bluesky — Dedicating my first post to this work and software, led by the incredibly meticulous and capable @fandingzhou.bsky.social! An earlier version of this was shared at the 2022 Bioconductor Conference (bioc2022.bioconductor.org/schedule/).
05.09.2025 13:32 — 👍 3 🔁 1 💬 1 📌 0

Gene expression changes aren’t just about mean shifts — variability shifts matter too, especially for aging. We're thrilled to introduce QRscore, a flexible non-parametric framework for detecting shifts in mean and variance across conditions. doi.org/10.1016/j.cr...
05.09.2025 02:15 — 👍 12 🔁 3 💬 1 📌 1
This work was led by my talented student Milind Jagota @milindjagota.bsky.social in collaboration with colleagues at UC Berkeley, UCSF (the Ye Lab @yimmieg.bsky.social), and Fred Hutch (the Matsen Lab @matsen.bsky.social). We are grateful to all co-authors for their enthusiasm and hard work. (n/n)
15.08.2025 13:17 — 👍 5 🔁 0 💬 0 📌 0
From a machine learning perspective, this work illustrates the value of high-quality negative examples. The paper is mostly focused on BCR light chains, but we are excited about extensions. (10/n)
15.08.2025 13:17 — 👍 2 🔁 0 💬 1 📌 0

We interpret what sequence features the model associates with dysfunction. One example is shown below. For a specific light chain V- and J- gene, we observe sharp selection on CDRL3 length, and on certain amino acids. (9/n)
15.08.2025 13:17 — 👍 1 🔁 0 💬 1 📌 0
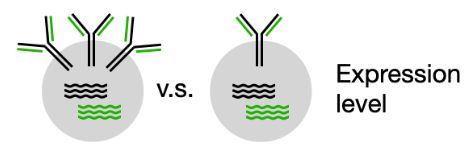

In new data, we find that very low scores are associated with reduced surface expression in naive B cells. To our knowledge, this is the first time expression variation in naive B cells has been linked to the light chain. (8/n)
15.08.2025 13:17 — 👍 1 🔁 0 💬 1 📌 0


B cells can further mutate antibodies to improve binding. We compare observed mutations to random control sets of mutations. Mutations that significantly decrease model scores appear to be selected out. However, this only works in a few positions. (7/n)
15.08.2025 13:17 — 👍 1 🔁 0 💬 1 📌 0


Models trained on allelic inclusion generalize to predict antibody properties with no direct training. Here we apply models to independent data measuring polyreactivity of human antibodies and observe correlation with polyreactivity. Baselines don’t capture this signal. (6/n)
15.08.2025 13:17 — 👍 1 🔁 0 💬 1 📌 0

We don’t know which sequence in each double-light B cell is “bad”, but we develop a training framework that doesn’t need this information. We compare with baseline approaches that don’t use the new allelic inclusion data. (5/n)
15.08.2025 13:17 — 👍 2 🔁 0 💬 1 📌 0

We propose using double-light B cells as negative examples for antibody machine learning. Double-light B cells can be observed at scale in some recent datasets of human antibodies. Each such cell has one “bad” sequence, whereas other cells all have functional antibodies. (4/n)
15.08.2025 13:17 — 👍 3 🔁 0 💬 1 📌 0

Most mature B cells express only the final, successful heavy and light chains (allelic exclusion). However, ~1% express two light chains (allelic inclusion). Previous work in mice has found that when this occurs, one of the light chains is dysfunctional. (3/n)
15.08.2025 13:17 — 👍 3 🔁 0 💬 1 📌 0

Natural antibodies are generated in B cells and tested for function (sufficient expression, low autoreactivity). If either the heavy or light chain fails, the B cell can try to generate it again. We usually can only sequence B cells that have passed all checkpoints. (2/n)
15.08.2025 13:17 — 👍 1 🔁 0 💬 1 📌 0
https://authors.elsevier.com/a/1lbX08YyDfuZWX
Antibodies are highly diverse, but most possible sequences are unstable or polyreactive. In this work, just published in Cell Syst., we propose a new source of data for modeling constraints from these properties. Our models show clear improvements in predicting Ab dysfunction. (1/n)
t.co/qCZERPUMPF
15.08.2025 13:17 — 👍 16 🔁 6 💬 1 📌 0


(1/4) 🧬 Why Sequence the Genomes of Earth’s Biodiversity?
The Earth BioGenome Project 🌍 is a global network of initiatives working together to create a complete genome library for all Eukaryotic life—from mushrooms 🍄 to mammals 🐘.
#biodiversity #genomes #sequence #earthbiogenome #education #stem
29.07.2025 20:59 — 👍 17 🔁 12 💬 1 📌 0

Germinal center clonal diversity trees as a musical score, a great image to start @victora.bsky.social's CCII seminar, "Replaying germinal center evolution on a quantified affinity landscape"
#GerminalCenter #Immunology
www.ccii.med.kyoto-u.ac.jp/en/event/the...
02.07.2025 02:42 — 👍 18 🔁 7 💬 1 📌 1
Associate Professor
DFCI & HMS
Professor, Molecular & Cellular Biology, Harvard University
Postdoc at Penn Genetics working in statistical genomics and computational biology.
Computational methods for epigenetic, CRISPR genome editing and single-cell genomics. Associate Professor at MGH / Harvard Medical School. http://pinellolab.org
Assistant Prof at D-BSSE, ETH Zurich, studying genetics of psychiatric disorders
www.nacailab.com
Bloomberg Distinguished Professor at Johns Hopkins University. http://schatz-lab.org
postdoc with Mark Daly at Broad Institute & MGH - curious about (large) chromosome alterations & (deep) human pedigrees + immunity, cancer & their interplay :)
Computational biologist, data scientist, digital artist | he, him | http://clauswilke.com/ | Opinions are my own and do not represent UT Austin.
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Genetics & Evolution, Faculty at Stanford & Freeman Hrabowski Scholar at HHMI
schumerlab.com
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
Virus-obsessed bioinformatician, DOE JGI Scientist, Enjoy exploring the viral world with #metagenomics and other cool #omics toys. He/him. Opinions my own.
Assistant prof at UCLA using the Bayes for the genomes https://pimentellab.com
Professor, Stanford University, Statistics and Mathematics. Opinions are my own.
Rapid evolutionary dynamics in viruses, cancer and bacteria. Assistant professor at UW Genome Sciences. federlab.github.io
Assistant Professor at UC Irvine. Transposons, epigenetics, and evolutionary genomics.
Associate Professor @ UC-Davis | Former Senior Director @ Exai Bio | Computational Biology, ML/AI, Genomics, RNA, Cancer
Professor, Investigator, mom, frequent flyer.
Using genome engineering to solve humanity’s greatest problems in health, climate & sustainable agriculture. UC Berkeley, UCSF, UC Davis. https://innovativegenomics.org/


























