
I’m in Oxford this Monday and Tuesday (morning). Looking forward to seeing many old friends and colleagues… and hopefully meet some new ones! talks.ox.ac.uk/talks/id/b6e...
02.11.2025 20:49 — 👍 7 🔁 2 💬 2 📌 0@sanejargon.bsky.social
Cancer and Ageing research; Pancreatic cancer transcriptomics; Bioinformatics; Long-read sequencing. CReST research fellow @ Cardiff University.

I’m in Oxford this Monday and Tuesday (morning). Looking forward to seeing many old friends and colleagues… and hopefully meet some new ones! talks.ox.ac.uk/talks/id/b6e...
02.11.2025 20:49 — 👍 7 🔁 2 💬 2 📌 0While I was taking a holiday last week, 2 super exciting preprints dropped, adding to another posted 6 days prior.
These papers describe a remarkable role for *recessive* variants in *RNU2-2* causing developmental and epileptic encephalopathy
🧵 by the amazing @christeldepienne.bsky.social 👇
1/3

I am super proud to present our new manuscript “Using SpliceAI to triage splice-altering variants in 7,220 individuals with rare conditions highlights limitations of the precomputed scores”
www.medrxiv.org/content/10.1...
Sept is approaching & I can already hear initial chats/enquiries for potential PhD studies happening!
If you're interested in a PhD, check out our department @cam.ac.uk www.phpc.cam.ac.uk/education-an...
We've a super interdisciplinary faculty spanning primary care, data science, epidemiology etc
🚨 New preprint led by amazing duo @rociorius.bsky.social and @alexblakes.bsky.social in collaboration with @cassimons.bsky.social, @dgmacarthur.bsky.social and many other amazing folks! ❤️
We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇
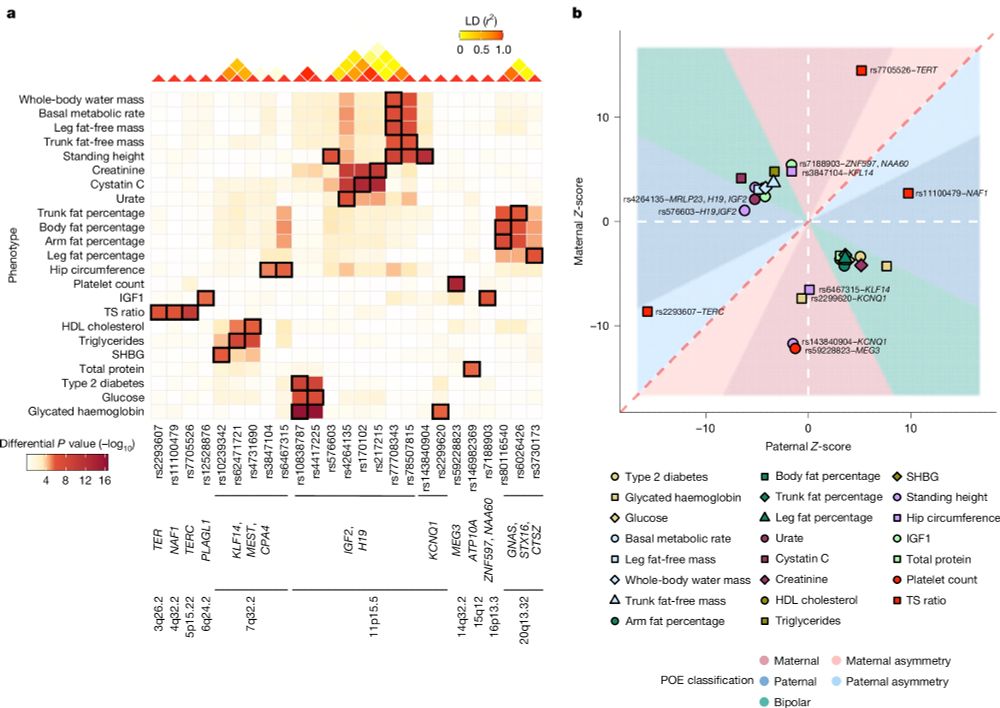
🚨 Our parent-of-origin study is out in Nature! 🧬
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
IMHO green computing & green software engineering are becoming even very important.
Check out our Green Algorithms Initiative for more tools and information www.green-algorithms.org
And the GREENER Principles which lay the foundation for a sustainability roadmap www.nature.com/articles/s43...
Our new draft V0.4 clonal/truncal somatic SV+CNV/CNA benchmark is at 42basepairs.com/browse/web/g.... Let us know if you have feedback to improve future versions!
21.07.2025 15:20 — 👍 1 🔁 1 💬 0 📌 0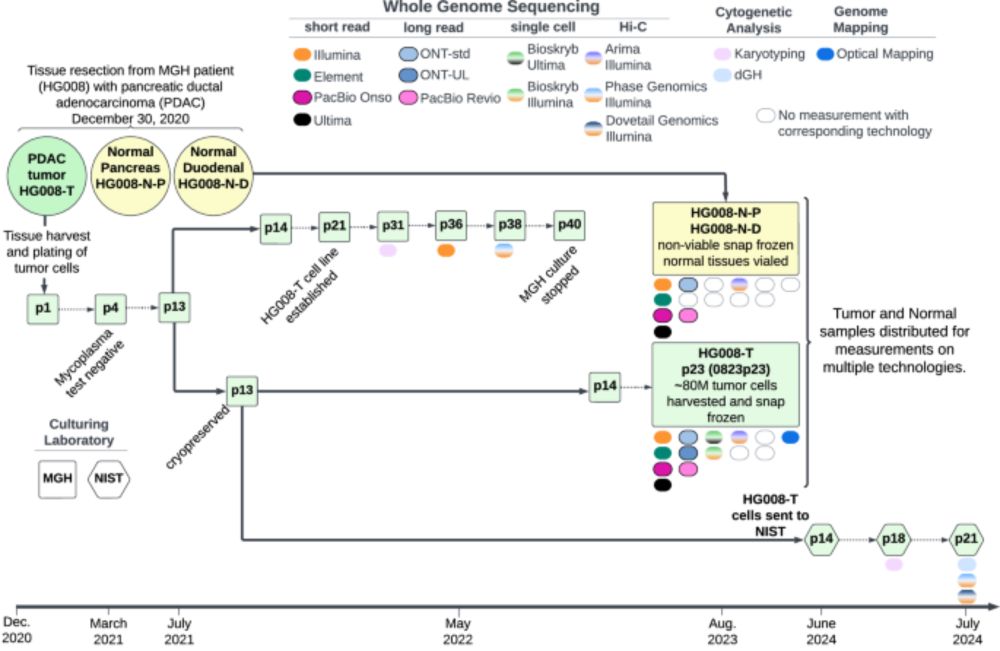
Our paper about extensive genomic data for a new broadly-consented pancreatic cancer cell line and matched normal tissue is now published at doi.org/10.1038/s415...! Stay tuned for draft GIAB benchmarks for somatic variants, and assemblies from these data coming soon! 1/2
21.07.2025 15:19 — 👍 3 🔁 1 💬 1 📌 0
Ydych chi ar LinkedIn? Rydym newydd lansio ein #cylchlythyr LinkedIn a fydd yn trafod #GwyddorauBywyd yng Nghymru!🧬✉️
Cael y diweddaraf am ddatblygiadau a straeon ysbrydoledig sy’n siapio dyfodol #arloesi ym maes #iechyd a #GofalCymdeithasol yng Nghymru a’r tu hwnt: www.linkedin.com/feed/update/...

Oeddech chi’n gwybod bod gennym ni gylchlythyr misol sy’n cynnwys y #newyddion diweddaraf ym maes #arloesi, yn ogystal â blogiau a digwyddiadau ar draws yr ecosystem #iechyd a #GofalCymdeithasol yng Nghymru a’r tu hwnt?
Tanysgrifiwch i Hwb Heddiw: lshubwales.com/cy/our-newsl...

Did you know we have a monthly #newsletter which features the latest #innovation news, blogs and events across the #health and #SocialCare ecosystem in #Wales and beyond?
Subscribe to Hub Highlights today: lshubwales.com/our-newsletter

Are you on LinkedIn? We’ve just launched our LinkedIn #newsletter dedicated to #LifeSciences in #Wales! 🧬✉️
Stay in the loop with developments, projects, and inspiring stories shaping the future of #health and #SocialCare #innovation across Wales and beyond: www.linkedin.com/feed/update/...
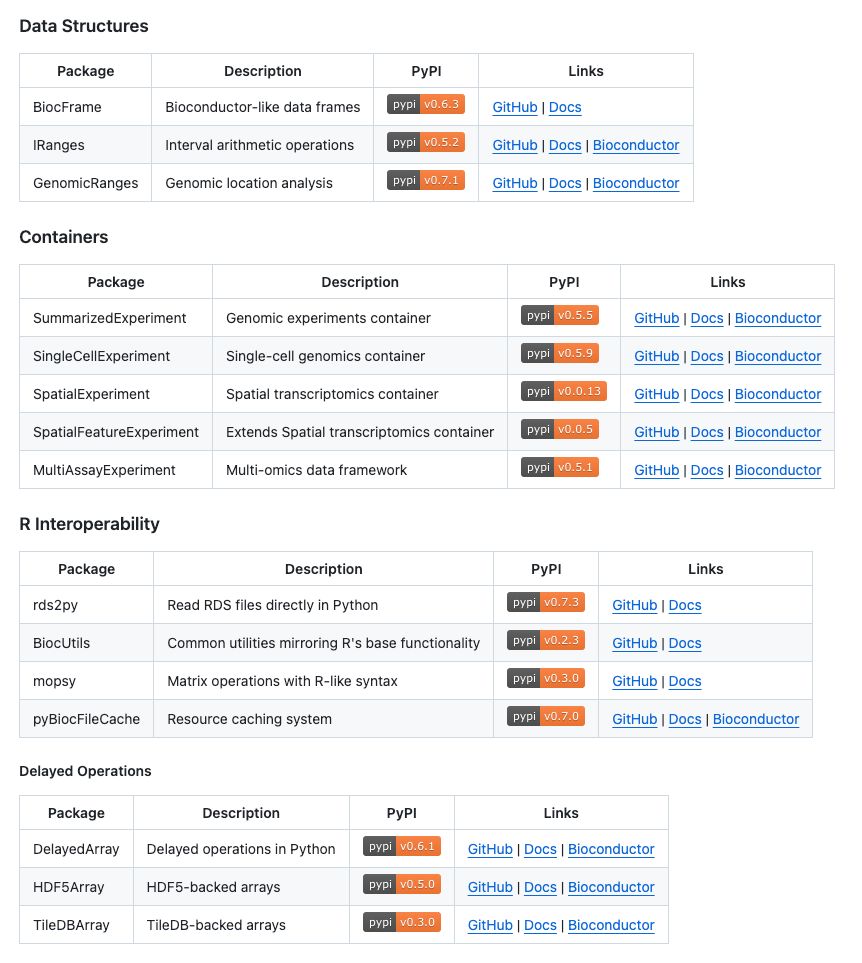
BiocPy brings Bioconductor's data structures and analysis tools to Python github.com/BiocPy #Rstats
Workshop materials: biocpy.github.io/BiocWorkshop...
Book: biocpy.github.io/tutorial/
Beta access to the hyper base calling models for Oxford @nanoporetech.com sequencing register.nanoporetech.com/hyper-baseca...
02.07.2025 19:21 — 👍 8 🔁 4 💬 0 📌 0
🚨 Our GW paper is out in Nature Methods!🥲
GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…
#Genomics #Bioinformatics

It's time!!!
An entire session of #eshg2025 on snRNA genes ❤️🤓
Join our new Wales Applied Virology Unit as a Lecturer in Infectious Disease Epidemiology / Behavioural Science!
See the advert here:
krb-sjobs.brassring.com/TGnewUI/Search…
And contact WAVU co-director David Gillespie for informal enquiries.
#NewPI #Lecturer #GroupLeader #Job #Opportunity

🚨I could not be more excited to share our new preprint on saturation genome editing of the small nuclear RNA (snRNA) RNU4-2:
www.medrxiv.org/content/10.1...
A super fun collaboration with incredible duo @gregfindlay.bsky.social @joachimdejonghe.bsky.social from @crick.ac.uk
🧬🖥️🩺
🧵1/12

Great to be visiting @nanoporetech.com as part of a cns Nanopore symposium meeting with pathologists and clinical scientists to discuss the application of nanopore sequencing in the clinic.
05.03.2025 10:51 — 👍 19 🔁 4 💬 0 📌 0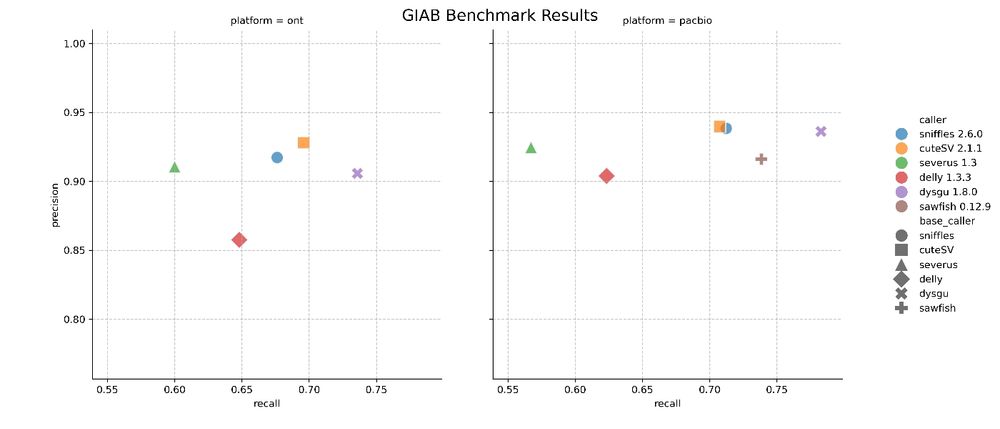
🧬🖥️ Just released Dysgu v1.8 which now supports phasing of structural variants when using long reads. Makes SV calling much more sensitive - see PacBio benchmark 👉 github.com/kcleal/SV_Be... dysgu repo github.com/kcleal/dysgu...
26.02.2025 11:37 — 👍 8 🔁 4 💬 0 📌 0
Minor-ish update to {ukbrapR} to v0.2.9 to fix 2 bugs and improve behaviour of some internal functions
github.com/lcpilling/uk...
Our first curated draft somatic structural variant benchmark for the new GIAB PDAC tumor cell line HG008-T is at ftp-trace.ncbi.nlm.nih.gov/ReferenceSam..., based on extensive short+long read sequencing data described in doi.org/10.1101/2024.... Feedback to improve future versions is very welcome!
20.12.2024 18:04 — 👍 15 🔁 8 💬 0 📌 0
🧬🖥️ Sharing an update to my SV benchmark repo. More callers have been added for the CMRG/GIAB benchmarks. github.com/kcleal/SV_Be.... Also now uses #Nextflow and tests the newest #PacBio Vega data, along with #ONT kit14. Results are on the GitHub page 👉
29.11.2024 13:40 — 👍 11 🔁 2 💬 2 📌 0This is worth looking at. Trying some genomes we have assembled with earlier versions of hifiasm, hifiasm plus herro and then this new version of hifiasm. The early assembly results suggest that now hifiasm alone is equivalent to what we got with herro but with less compute!
28.11.2024 12:58 — 👍 21 🔁 5 💬 0 📌 0I know many of you have been awaiting us launching transcript expression data in gnomAD. We were waiting for the GTEx v10 release which is now out so we are finally able to launch this. Enjoy!!
22.11.2024 14:44 — 👍 53 🔁 20 💬 1 📌 1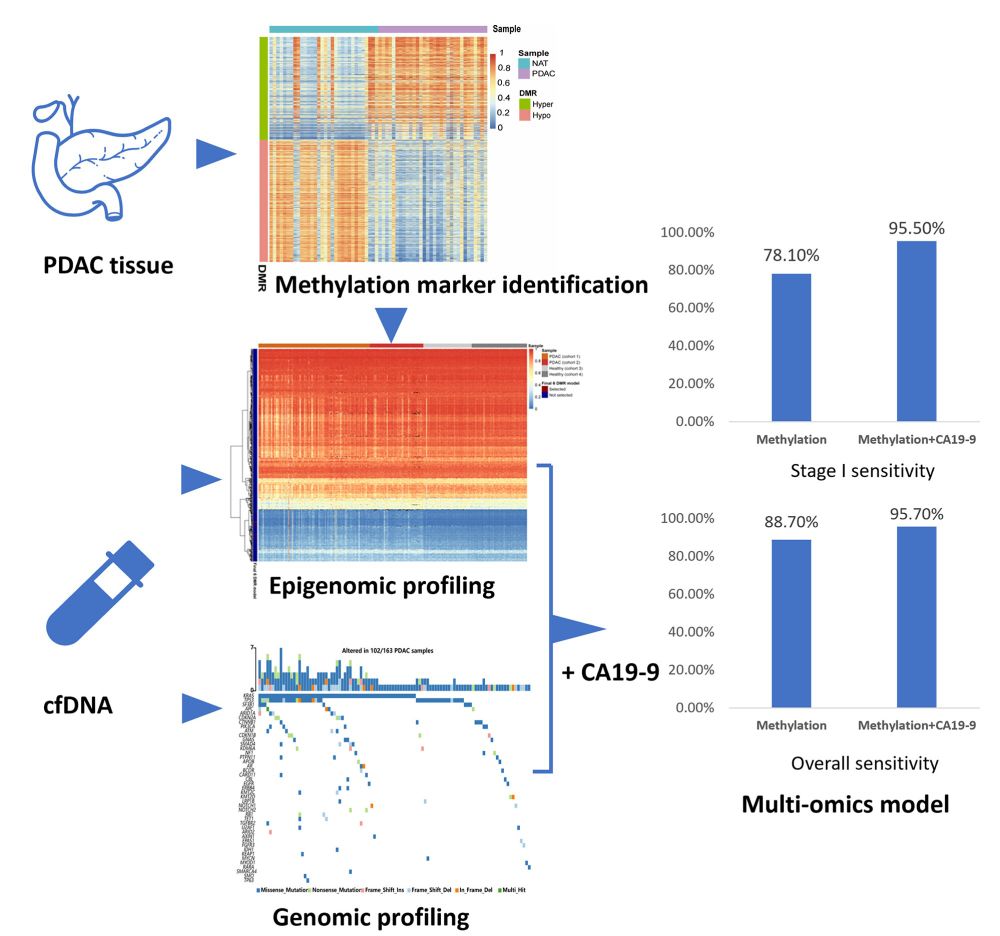
Circulating cell-free DNA methylation-based multi-omics analysis allows early diagnosis of pancreatic ductal adenocarcinoma #PancreaticCancer
febs.onlinelibrary.wiley.com/doi/10.1002/...
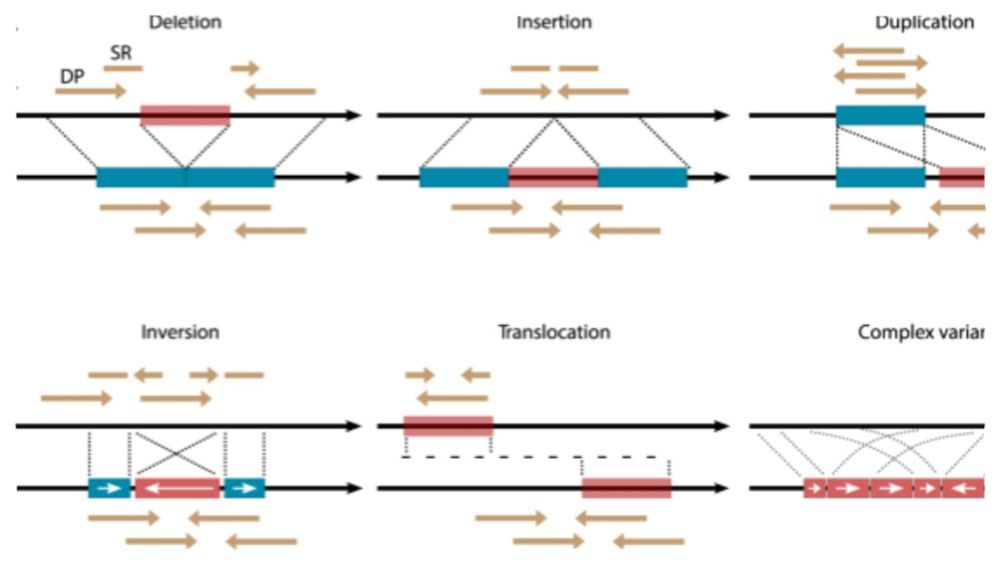
Interested in Structural Variants?
Join for this 3-day course with @sedlazeck.bsky.social
and Luis Paulin in December (2-4): physalia-courses.org/courses-work...
We’ll cover tools and workflows for SVs using @illumina
@oxfordnanopore.bsky.social and @pacbio.bsky.social data!
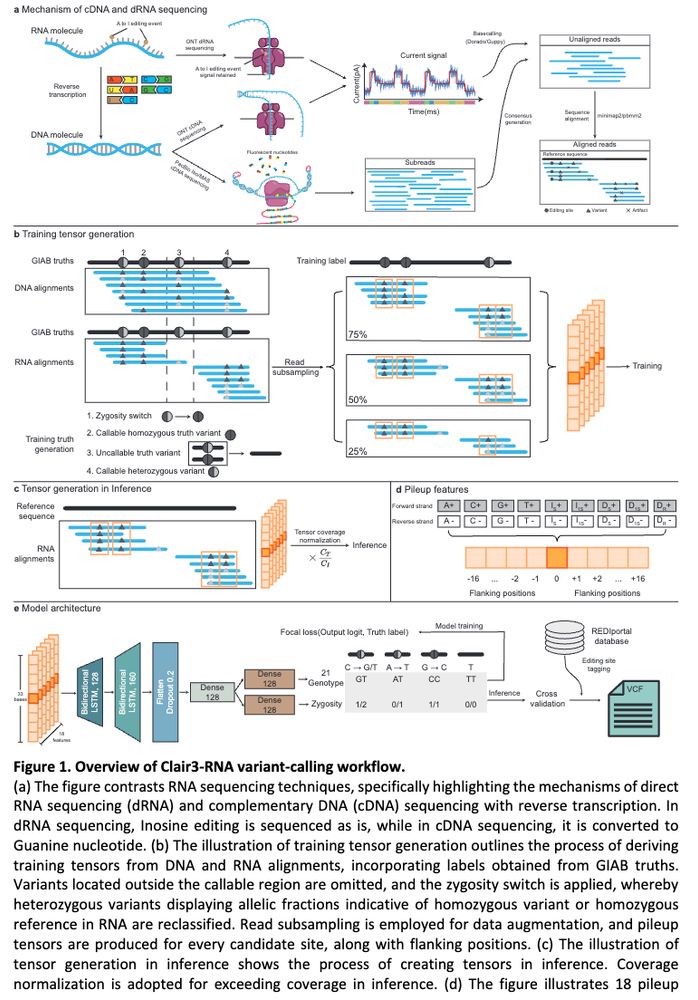
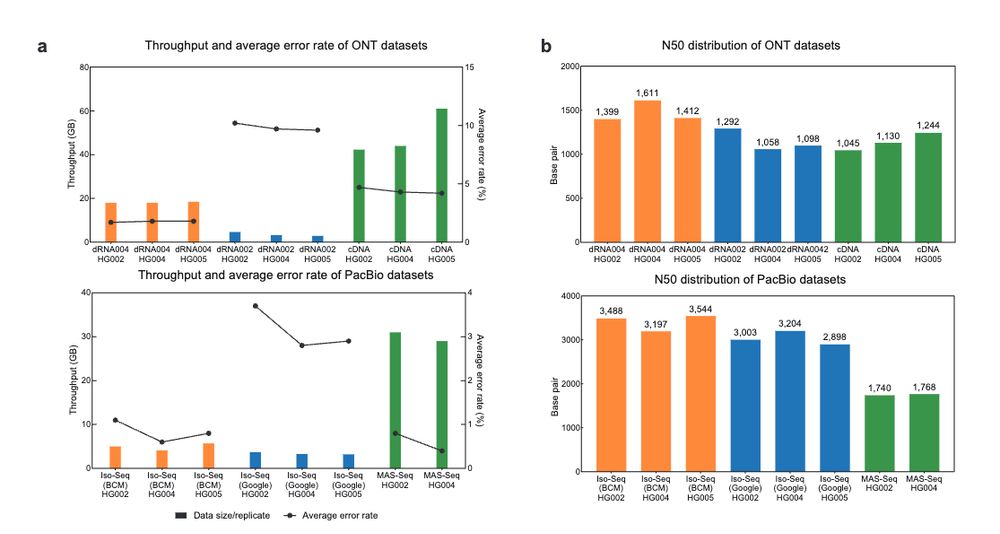
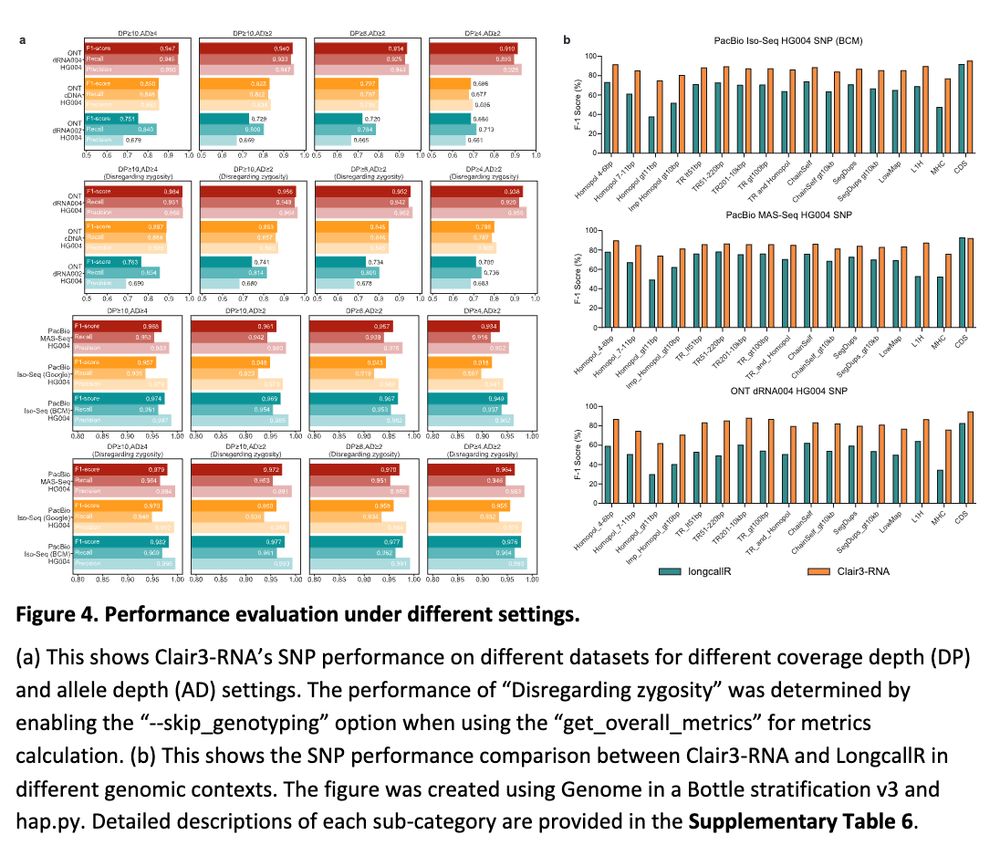
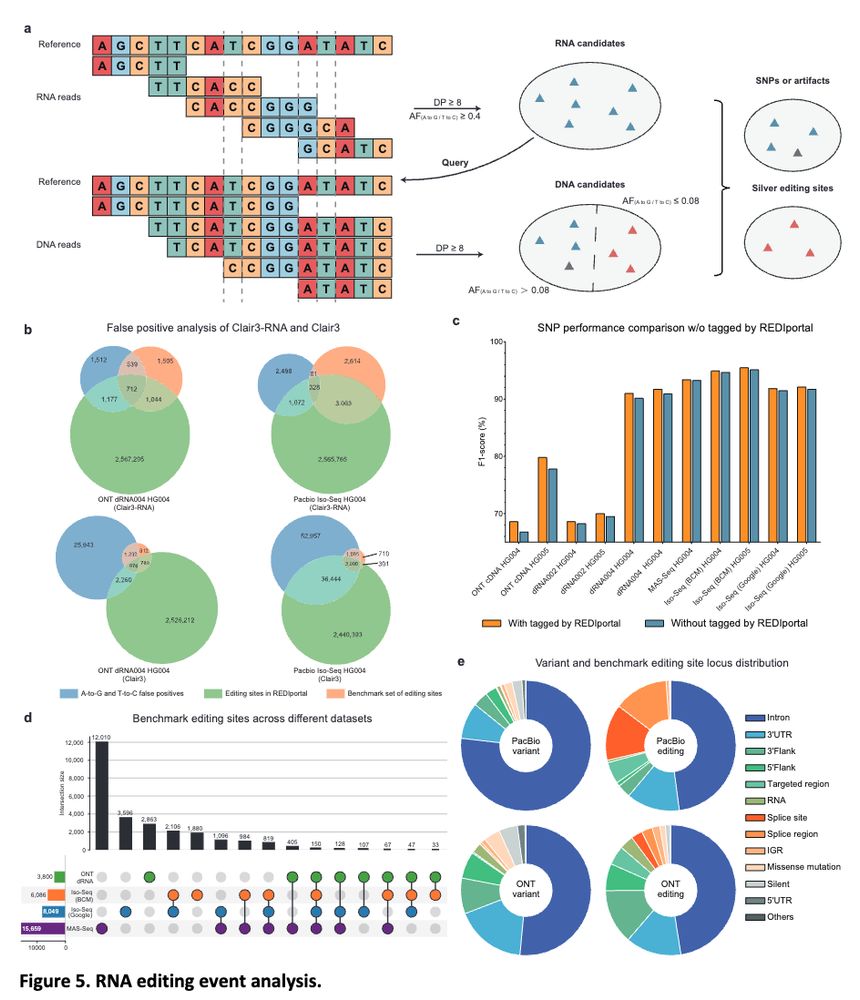
Clair3-RNA: A deep learning-based small variant caller for long-read RNA sequencing data https://www.biorxiv.org/content/10.1101/2024.11.17.624050v1 🧬🖥️🧪 https://github.com/HKU-BAL/Clair3-RNA
20.11.2024 16:30 — 👍 11 🔁 3 💬 0 📌 0
New interval lookup library just hit for Rust, C++ and Python — claims best in class performance! Pretty cool : github.com/kcleal/super...
20.11.2024 15:18 — 👍 42 🔁 9 💬 4 📌 0