This is the kind of studies I like to see. Great job 👍
17.02.2026 11:16 — 👍 1 🔁 0 💬 0 📌 0Daniel Estévez Barcia
@daeb-genes.bsky.social
Population genetics, molecular ecology, sex evolution, marine biology. Greenland Institute of Natural Resources
@daeb-genes.bsky.social
Population genetics, molecular ecology, sex evolution, marine biology. Greenland Institute of Natural Resources
This is the kind of studies I like to see. Great job 👍
17.02.2026 11:16 — 👍 1 🔁 0 💬 0 📌 0Conservation genomics and assisted introgression is saving the American chestnut from extermination by invasive non-native pathogens. #consgen
13.02.2026 20:48 — 👍 16 🔁 6 💬 2 📌 0Awesome study!
11.11.2025 21:53 — 👍 1 🔁 0 💬 1 📌 0Thought exactly the same when I read the title of the post
03.11.2025 16:52 — 👍 2 🔁 0 💬 0 📌 0
📣📣📣 CONSERVATION GENETICS WORKSHOP -
Few more spaces left!
ConGen Population Genomic Data Analysis Course
🧬💻💡
University of Pretoria, South Africa 🌍
7-14 December + field trip to Kruger NP!
🦁🐃🐘🐆🦏
More info & registration here: www.umt.edu/congen/africa/

Happy to see my former @nbis.se advisory program student Zach Nolen's new pipeline PopGLen for analyses of short-read data using genotype-likelihood based methods out!
Github: github.com/zjnolen/PopG...
Paper: academic.oup.com/bioinformati...
Neat job alert 🧬🐺🦌
Montana Fish Wildlife & Parks is hiring a wildlife conservation geneticist based in Missoula, MT. The position is joint with @umontana.bsky.social, working with Dr. Ryan Kovach on lots of neat wildlife #consgen projects. Reach out to Ryan with questions.
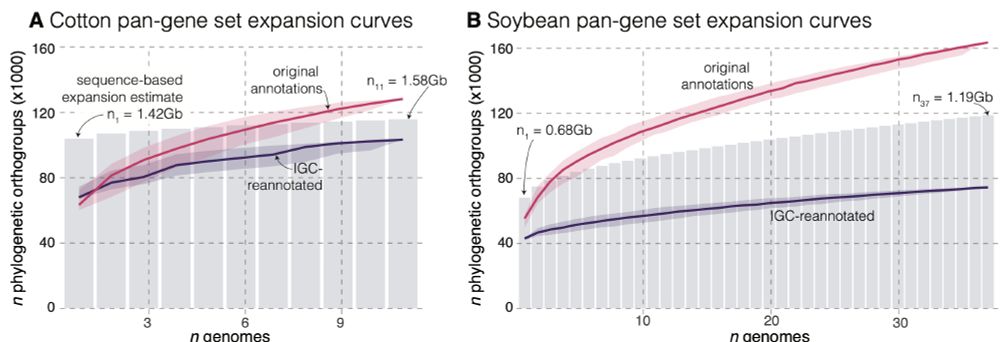
pangenome 'expansion' curves for cotton and soybean
Determining presence-absence variation (PAV) across reference genomes is a major goal of pangenome analysis. It turns out that A LOT of gene PAV is due to methodological artifacts.
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...

Go check out our new post on the Methods Blog!
Find out how a simple dust cloth might revolutionize airborne eDNA monitoring 🧬 🌍 🧪
buff.ly/BhIKcKq

Interested in how new sex chromosomes evolve in birds? Our NSF-funded collaborative work with the (Daven) Presgraves lab at @urochester.bsky.social is out this week in @pnas.org. Check it out!
www.pnas.org/doi/10.1073/...
Aah ok. But just as reminder, the original post was between NovaseqX and Novaseq6000 ☺️
22.05.2025 16:52 — 👍 1 🔁 0 💬 0 📌 0I believe both technologies use a two colour chemistry, and they both tend to have an increase in Poly-G
22.05.2025 16:31 — 👍 0 🔁 0 💬 1 📌 0Let me know if you manage, having struggles myself...
06.05.2025 13:09 — 👍 0 🔁 0 💬 1 📌 0Help! Is anyone else finding batch effects between whole genome sequences generated with Illumina NovaSeq6000 vs NovaSeqX?
Any suggestions or solutions would be hugely appreciated! 🙏
#PopGen #ConGen #Bioinformatics #Genomics 🧬🖥️🧪
Downsize reads depth, and also use the option in AGSD to make PCA instead of using PCAngsd
06.05.2025 12:53 — 👍 0 🔁 0 💬 1 📌 0Yup, I have found it. I am now inspecting bamqc in my different batches. I have a few things considered: poly-G removal with improved code in fastP (done but does not remove the batch effect)
06.05.2025 12:53 — 👍 0 🔁 0 💬 2 📌 0Still ongoing. I have however encountered a big issue in the form of batch effects and have suffered some drawbacks from poor tissue quality. Bioinformatic corrections exist, but so many headaches could be afforded with good standard fieldwork for genetic work
29.04.2025 14:43 — 👍 2 🔁 0 💬 0 📌 0
Past conservation efforts reveal which actions lead to positive outcomes for species journals.plos.org/plosbiology/...
28.04.2025 18:30 — 👍 6 🔁 3 💬 0 📌 0
#consgen authority Fred Allendorf cries out in despair for American science in the Missoulian newspaper. It’s terrifying to watch this destruction unfold day by day. My heart goes out to my American colleagues and friends. Stay strong and fight.
10.04.2025 05:38 — 👍 25 🔁 15 💬 1 📌 1