Original post on social.edu.nl
new blog post: "Rescuing @wdscholia #2: getting closer" https://chem-bla-ics.linkedchemistry.info/2025/12/31/rescuing-scholia-2-getting-close.html https://doi.org/10.59350/6t2qh-2f839
"But we are getting close. So, please give qlever.scholia.wiki a go, and let us know your observations. As […]
31.12.2025 11:14 — 👍 2 🔁 5 💬 0 📌 0

MIBiG 5.0 Annotathons Registration Form
Planned for spring 2026
Speaking of MIBiG hackathons: iteration 5.0 it is planned for spring 2026 and registrations are open! Please sign up if you are interested and feel free to share with your colleagues! (7/8) forms.gle/JKsf3XL3d1XN...
11.12.2025 19:19 — 👍 4 🔁 2 💬 1 📌 2

Strategies for community-sourced biocuration in bioinformatics: a case study on MIBiG 4.0
Abstract. Biocuration is essential to transform molecular sequence data into standardized, machine-readable resources. Such curated datasets enable compara
Have you ever used a #bioinformatics #database and were frustrated by its lack of coverage? Did you ever think about starting your own resource? We just published a new strategy for community-driven #biocuration, based on our experiences with the #MIBiG database (1/8)! doi.org/10.1093/bib/...
11.12.2025 19:19 — 👍 38 🔁 16 💬 1 📌 0
👉 Point your friends, family and connections to our new video 📹 on the point of bioinformatics!
The 90-second video is available in:
🇬🇧 youtu.be/yjbv1WZcM2M
🇫🇷 youtu.be/BG-h_nxwSE4
🇩🇪 youtu.be/e4S4m1IhfiU
🇮🇹 youtu.be/V4QcZHjnsGo
10.12.2025 08:55 — 👍 11 🔁 6 💬 0 📌 0

A Perspective on Unintentional Fragments and Their Impact on the Dark Metabolome, Untargeted Profiling, Molecular Networking, Public Data, and Repository Scale Analysis
In/postsource fragments (ISFs) arise during electrospray ionization or ion transfer in mass spectrometry when molecular bonds break, generating ions that can complicate data interpretation. Although ISFs have been recognized for decades, their contribution to untargeted metabolomics─particularly in the context of the so-called “dark matter” (unannotated MS or MS/MS spectra) and the “dark metabolome” (unannotated molecules)─remains unsettled. This ongoing debate reflects a central tension: while some caution against overinterpreting unidentified signals lacking biological evidence, others argue that dismissing them too quickly risks overlooking genuine molecular discoveries. These discussions also raise a deeper question: what exactly should be considered part of the metabolome? As metabolomics advances toward large-scale data mining and high-throughput computational analysis, resolving these conceptual and methodological ambiguities has become essential. In this perspective, we propose a refined definition of the “dark metabolome” and present a systematic overview of ISFs and related ion forms, including adducts and multimers. We examine their impact on metabolite annotation, experimental design, statistical analysis, computational workflows, and repository-scale data mining. Finally, we provide practical recommendations─including a set of dos and do nots for researchers and reviewers─and discuss the broader implications of ISFs for how the field explores unknown molecular space. By embracing a more nuanced understanding of ISFs, metabolomics can achieve greater rigor, reduce misinterpretation, and unlock new opportunities for discovery.
The existence of unintentional fragmentation (often referred to as in-source fragments) in untargeted #metabolomics data can cause uncertainty among newcomers to the field and skepticism among data consumers such as medical experts or biologists.
pubs.acs.org/doi/10.1021/...
03.12.2025 15:29 — 👍 19 🔁 6 💬 1 📌 1

Reliable data for better care
Every medical revolution has been driven by a technological breakthrough: chemical synthesis paved the way for modern medicines, genetics for the first gene therapies. The next frontier will be determ...
💡 No AI in health without great data. In @BilanMagazine’s health supplement, Christophe Dessimoz @dessimoz.bsky.social makes the case for Switzerland as a leader in trusted, high-quality health data — and how turning data into public value starts with strong infrastructures.
👇 Read more
30.10.2025 09:12 — 👍 9 🔁 4 💬 0 📌 1
Available to read now via Open Access:
pubs.rsc.org/en/content/a...
Fast, general-purpose metabolome analysis by mixed-mode liquid chromatography–mass spectrometry
Nicola Zamboni et al @nzamboni.bsky.social @alaaothman.bsky.social @ethz.ch
#MassSpec
30.10.2025 09:28 — 👍 2 🔁 4 💬 0 📌 0

Ilse Falls in the Harz National Park, Germany. Text says: Wikidata is now the second Wikimedia project recognized as a digital public good.
Wikidata has been recognized as a digital public good by the Digital Public Goods Alliance (DPGA) 🎉
Learn how Wikidata, which houses over 1.6 billion facts, advances education, innovation, and public institutions ➡️ wikimediafoundation.org/news/2025/10...
29.10.2025 18:24 — 👍 136 🔁 36 💬 0 📌 0

Wikidataians celebrate Wikidata being recognised as a Digital Public Good
Every day, thousands of editors on #Wikidata provide open, reliable and trustworthy data to the world helping make technology more open and inclusive.
Today, we are thrilled to announce that Wikidata has been recognized by the @DPGAlliance as a #DigitalPublicGood.
29.10.2025 18:21 — 👍 19 🔁 8 💬 1 📌 2

Correspondence in Nature: to make FAIR a reality, fund the data resources and expertise—not just data-management plans. A call to action co-signed with @francesarnold.bsky.social, Rich Roberts, and Tim Hubbard. doi.org/10.1038/d415...
23.10.2025 06:26 — 👍 7 🔁 1 💬 1 📌 1
@skepteis.bsky.social brought us to a local place close to Arnhem! 🥞
15.10.2025 07:17 — 👍 2 🔁 0 💬 1 📌 0
Best part was the quadruple, and now also the Pannenkoeks!
15.10.2025 05:05 — 👍 1 🔁 0 💬 1 📌 0
Reminder 👇
Interested in improving our R tools for #MassSpectrometry data analysis and integrating them into Galaxy?
⏲️ 3 year position
📍 Bolzano, 🇮🇹
👉 apply if you like:
- #rstats SW development
- @bioconductor.bsky.social
- large-scale #metabolomics data analysis
- hiking ⛰️
🔗 bit.ly/46AMawx
06.10.2025 14:29 — 👍 7 🔁 6 💬 1 📌 0
Original post on mastodon.social
I still love the @wdscholia DOI redirection tool. When the DOI is not listed in @wikidata, it will use @larsgw's @citationjs to create QuickStatements that can be used with @magnusmanske's tool to create a new Wikidata item.
And it supports ORCID to link to authors, "title in HTML", mul, and […]
27.09.2025 09:12 — 👍 2 🔁 1 💬 0 📌 0
Aaand it's out! Meet MITE - the natural product tailoring enzyme database, just published in @narjournal.bsky.social! MITE DB captures the substrate- and reaction-specificity of tailoring enzymes, allowing to capture this information in a human- and machine-readable way! doi.org/10.1093/nar/...
27.09.2025 09:11 — 👍 46 🔁 18 💬 1 📌 2
@mehdibeni.bsky.social drove it forward, and it was a pleasure to contribute with @robinschmid.bsky.social, @Wout Bittremieux, @Olivier Cailloux, and @jjjvanderhooft.bsky.social.
It places the scalability of MS-based metabolomics for natural extracts as a central challenge rather than a side issue.
21.09.2025 15:55 — 👍 4 🔁 4 💬 0 📌 0

A Perspective on Unintentional Fragments and their Impact on the Dark Metabolome, Untargeted Profiling, Molecular Networking, Public Data, and Repository Scale Analysis.
In/post-source fragments (ISFs) arise during electrospray ionization or ion transfer in mass spectrometry when molecular bonds break, generating ions that can complicate data interpretation. Although ISFs have been recognized for decades, their contribution to untargeted metabolomics - particularly in the context of the so-called “dark matter” (unannotated MS or MS/MS spectra) and the “dark metabolome” (unannotated molecules) - remains unsettled. This ongoing debate reflects a central tension: while some caution against overinterpreting unidentified signals lacking biological evidence, others argue that dismissing them too quickly risks overlooking genuine molecular discoveries. These discussions also raise a deeper question: what exactly should be considered part of the metabolome? As metabolomics advances toward large-scale data mining and high-throughput computational analysis, resolving these conceptual and methodological ambiguities has become essential. In this perspective, we propose a refined definition of the “dark metabolome” and present a systematic overview of ISFs and related ion forms, including adducts and multimers. We examine their impact on metabolite annotation, experimental design, statistical analysis, computational workflows, and repository-scale data mining. Finally, we provide practical recommendations - including a set of dos and don’ts for researchers and reviewers - and discuss the broader implications of ISFs for how the field explores unknown molecular space. By embracing a more nuanced understanding of ISFs, metabolomics can achieve greater rigor, reduce misinterpretation, and unlock new opportunities for discovery.
If you’ve been following #metabolomics literature, you’ve probably seen a lot of debate on in-source fragmentation. We’ve put together a manuscript to clarify what it is, how to deal with it, and what it means for discovery in #metabolomics and #exposomics.
doi.org/10.26434/che...
23.08.2025 02:31 — 👍 4 🔁 3 💬 0 📌 0

Into natural product biosynthesis & tailoring enzymes? Frustrated by the lack of a dedicated resource to explore their functions? Tired of endless literature searches for reaction info? Meet the MITE database, freely available at mite.bioinformatics.nl. Preprint: doi.org/10.26434/che... (1/8)
21.08.2025 10:52 — 👍 36 🔁 17 💬 1 📌 3
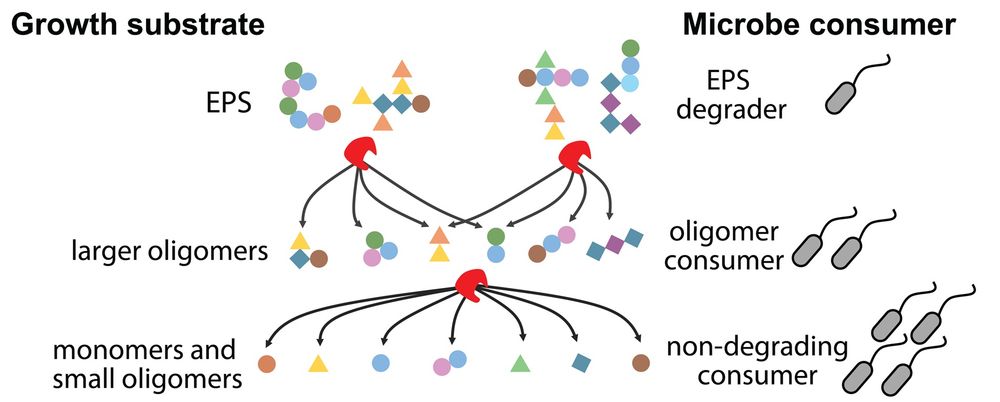
Graphical illustration of how EPS is enzymatically degraded in multiple steps into smaller fragments, fueling the growth of non-degrading species: EPS degraders break down EPS into larger oligomers for oligomer consumers; further degradation into monomers and small oligomers supports non-degrading consumers.
What is the role of extracellular polymeric substances (EPS) in carbon exchange among microbial species? @sammy-pontrelli.bsky.social &co show that #EPS, formed via #chitin degradation, drives #MicrobialDiversity by acting as a sequentially degraded #CarbonSource @plosbiology.org 🧪 plos.io/3J9kbuu
31.07.2025 09:01 — 👍 21 🔁 6 💬 1 📌 0
Original post on social.edu.nl
heading tomorrow to the InChI Technical Exchange Meeting Summer 2025 in Aachen/DE
Looking forward to it, and particularly talking about the InChI for inorganics and trying that in @wikidata :) See https://doi.org/10.26434/chemrxiv-2025-53n0w
And also the nano InChI, see […]
20.07.2025 19:39 — 👍 5 🔁 6 💬 1 📌 0
@skepteis.bsky.social You're not alone 😉
18.07.2025 11:47 — 👍 1 🔁 0 💬 0 📌 0

Funny feed showing Daniel Probst and ChemDraw both turning 40
My feed just made it even better:
As old as ChemDraw 🥳
18.07.2025 11:46 — 👍 1 🔁 0 💬 1 📌 0
🚀 We’ve launched the new MassBank! Now live at massbank.eu & massbank.jp — redesigned with a faster backend, better search, and powerful tools for exploring & sharing mass spectral data. Enjoy the fresh experience! Feedback and ideas welcome, please post them on github.com/MassBank/Mas...
16.07.2025 18:57 — 👍 14 🔁 5 💬 0 📌 0
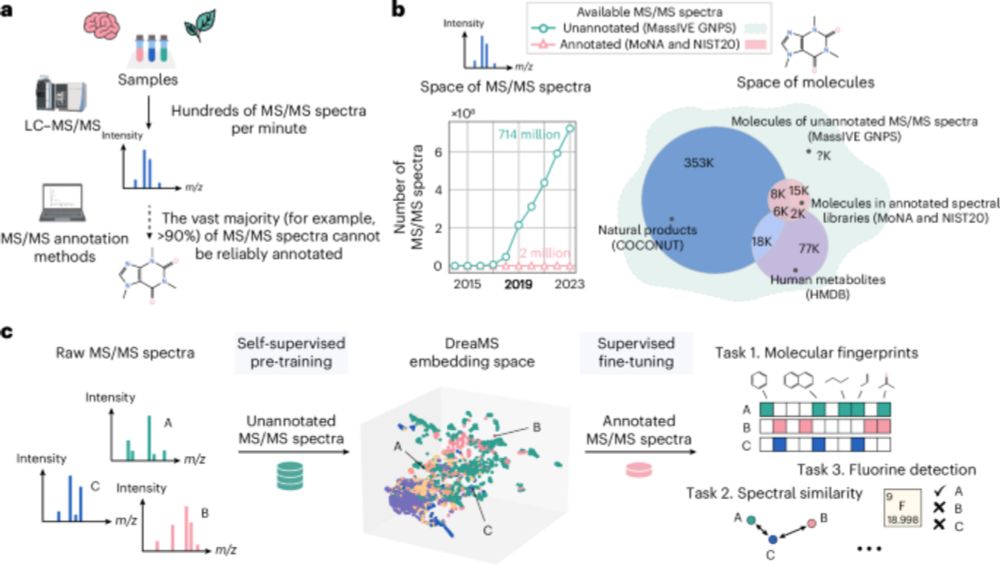
Self-supervised learning of molecular representations from millions of tandem mass spectra using DreaMS - Nature Biotechnology
A transformer model is used to construct the DreaMS Atlas—a molecular network of 201 million MS/MS spectra.
Mass spectrometry is a key method to discover and identify molecules in biological and environmental samples. Yet, >90% of mass spectra remain hard to interpret. In our recent paper, we present DreaMS — a foundation model to interpret mass spectra of small molecules.
www.nature.com/articles/s41...
26.05.2025 13:44 — 👍 25 🔁 10 💬 1 📌 0

Excited to welcome our new Agilent Revident Q-TOF! We’re developing new approaches combining mass spectrometry and enzyme assays to study microbial interactions and carbon sequestration. We're hiring postdocs — reach out if interested!
pontrellilab.sites.vib.be/en
28.04.2025 11:17 — 👍 4 🔁 3 💬 1 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
Research engineer position on methods and tools for the construction, maintenance and querying of a decentralized knowledge hub in metabolomics
lnkd.in/erh_eeTy
31.03.2025 14:08 — 👍 0 🔁 2 💬 0 📌 0
Metabolomics, Mass spectrometry informatics | Postdoc at Dorrestein Lab, UCSD
The home of premier fundamental discoveries, inventions and applications in the analytical and bioanalytical sciences.
Published by @rsc.org 🌐 Website: rsc.li/analyst-rsc
Dixi et salvavi animam meam.
Post-doc at Dorrestein lab. Interested in all things metabolism, microbes and enzymes.
Postdoctoral fellow NSERC/FRQS1 at UC San Diego in the Dorrestein Lab | Raffatellu Lab
Microbiology | Bacterial molecular genetics | Untargeted metabolomics
Connecting research and researchers. https://linktr.ee/orcid_org
#metabolomics and #lipidomics scientist - University of Geneva 🇨🇭
Advancing untargeted LC-MS-based approaches for toxicological applications
Tropical Plant Diversity & Ethnobotany
Professor at University of Zurich🇨🇭
🍃Dad of two🍃 www.rcamaraleret.com
Natural Products Chemist
Interested in metabolomics and proteomics
Molecular Identifier
https://www.inchi-trust.org and https://www.iupac.org
He/Him. Evolution and Bioinformatics. Posts mine alone, in English (mostly) & French. Chair of @dee-unil.bsky.social, Prof at @fbm-unil.bsky.social, group leader at @sib.swiss. PI of @bgee.org
🦣 Main account: https://ecoevo.social/@marcrr
Associate Prof. University of Pittsburgh. Computational and Systems Biology. Mechanisms of Evolutionary Innovation. Yeast. www.carvunis.com.
Professor of Genome Sciences University of Washington, Seattle. Interested in proteomics and mass spectrometry.
Europäischer Bündner, Nationalrat, Vizepräsident SP Schweiz, Präsident Schweizerische Gesellschaft für Aussenpolitik SGA|ASPE
A happy (and a bit stressed) PhD student, doing her best to navigate software development in R and having fun doing it.
Wanna learn LC-MS/MS analysis in R ? Click https://github.com/rformassspectrometry/Metabonaut
Assistant Professor and Group Leader
KU Leuven Biology, Division of Biotech of Plants and Microbes
VIB Center for Microbiology
Nonprofit advancing discovery through rapid, free sharing of scientific communications. New home of bioRxiv and medRxiv.















