tl;dr: populations encoding 2- and 3-D quantities need shape and size heterogeneity to increase encoded information. We show how one can measure these heterogeneities using Bayesian inference, which offers a principled way of handling noisy, incomplete neural data. [n=16/n]
31.07.2025 18:02 — 👍 3 🔁 0 💬 0 📌 0
More broadly, this framework produces a bunch of testable predictions beyond place fields. And, we propose a principled method for testing these predictions in other populations. [15/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
With our theory, we can also quantify just how much more information about spatial position these populations encode than if they all shared the same size/shape. [14/n]
31.07.2025 18:02 — 👍 1 🔁 0 💬 1 📌 0
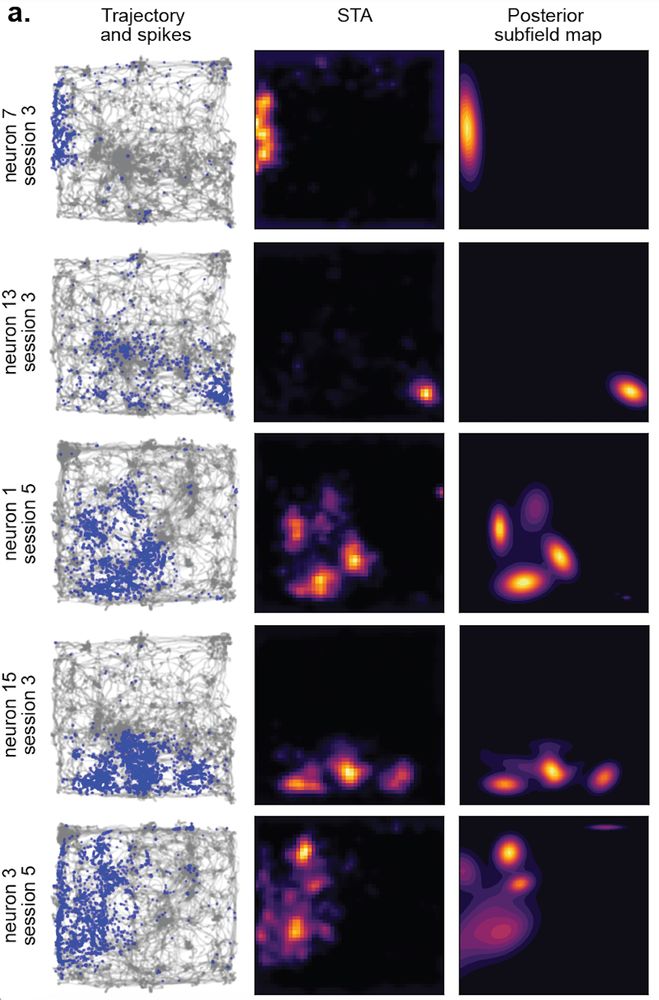
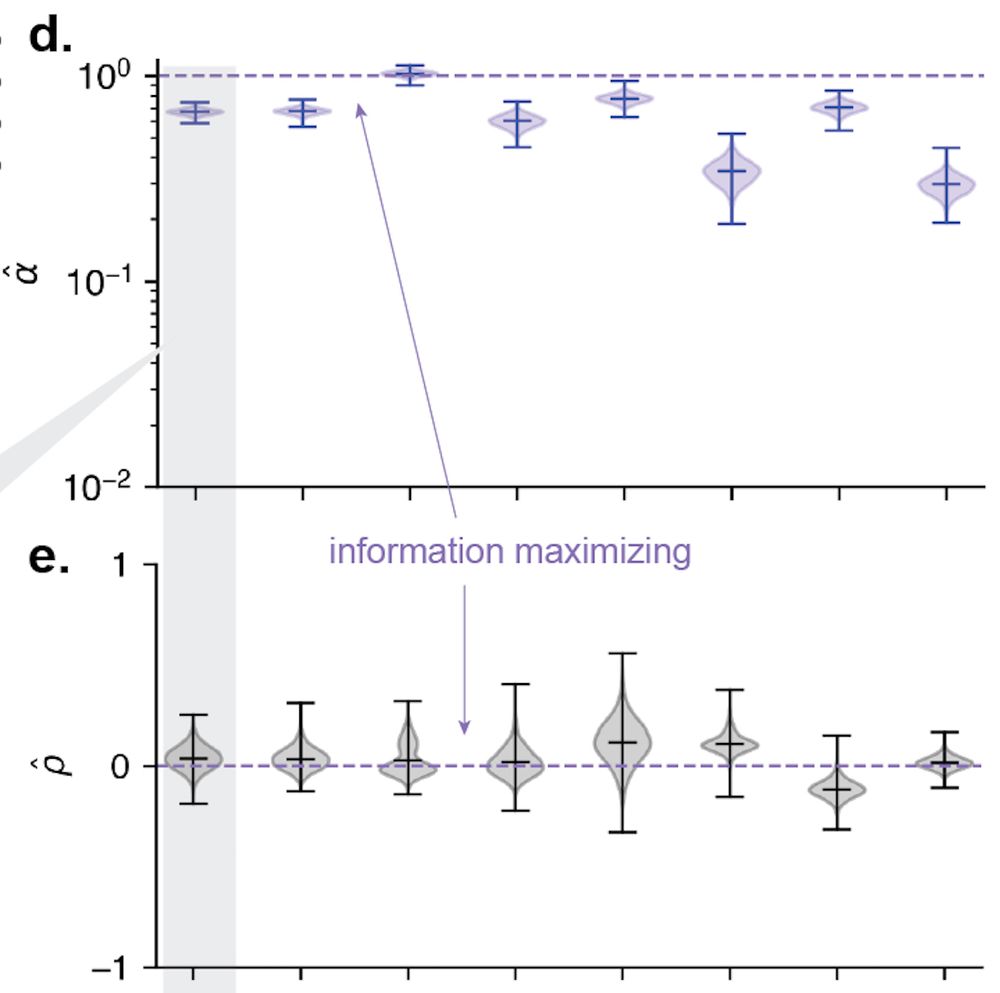
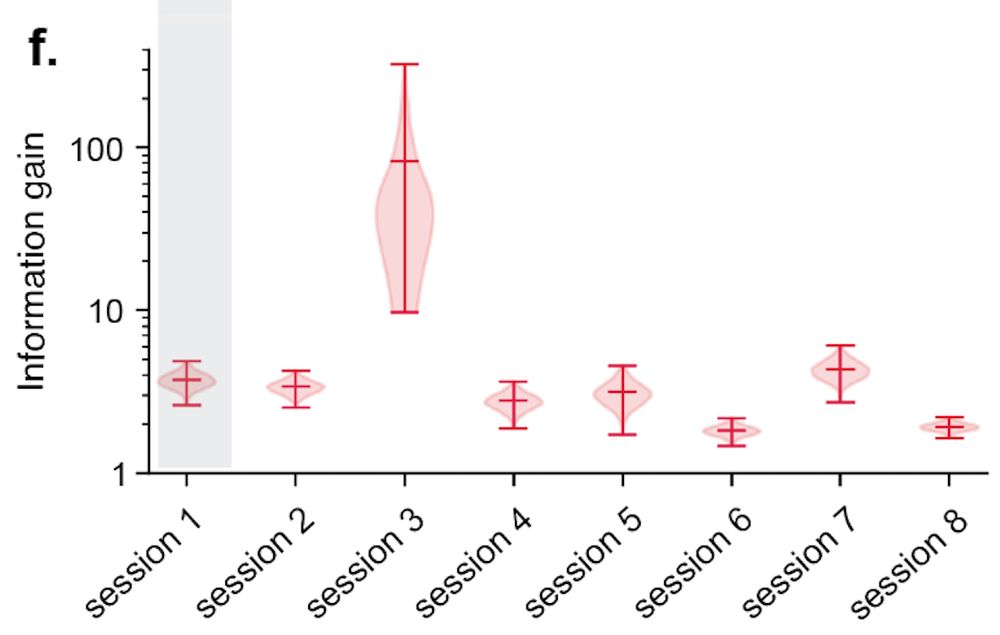
With our new method in hand, we turned back to rodent place fields. As the theory predicts, place fields appear to exhibit high degrees of size and shape heterogeneity! (ticks in d,e are sessions) [13/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
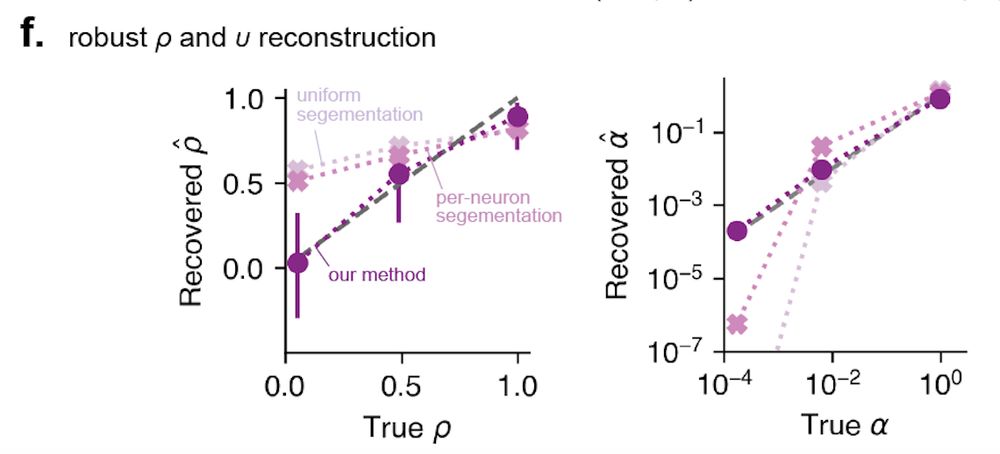
We show that doing this overcomes the issues we identified with tuning measurement based on the STA. With it, we can identify shape and size heterogeneity in simulated populations far more accurately. Our method is also less biased by the occupancy pattern of the animal. [12/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
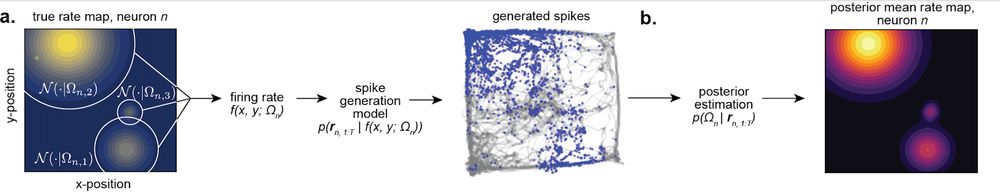
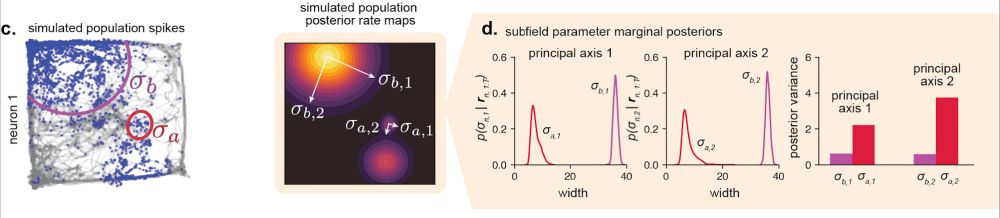
We propose a new Bayesian approach. We use it to explicitly quantify how uncertain we are about a field’s tuning based on how limited data is. Then we use this uncertainty to weight our estimates of field properties when measuring the degrees of shape and size heterogeneity. [11/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
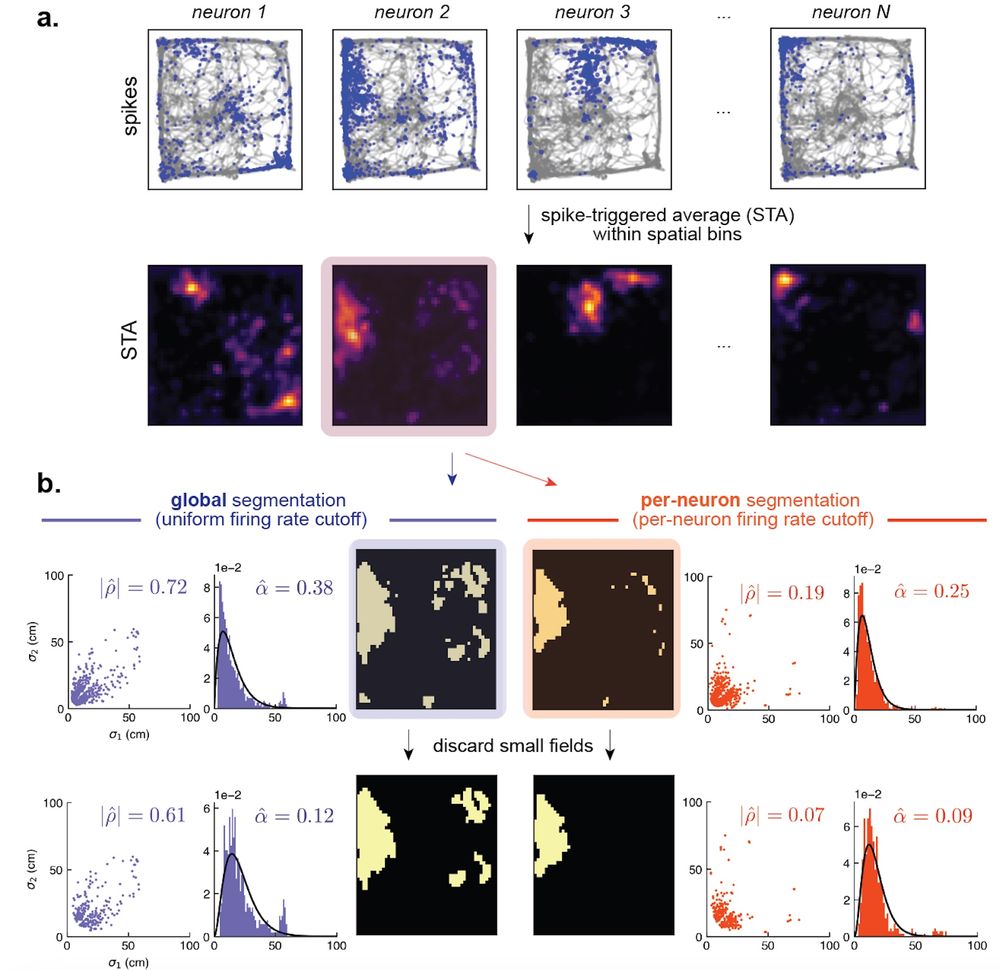
In the context of place cells, this refers to the rat not traversing the environment perfectly uniformly. These biases can skew the measurements of shape and size heterogeneity. So, we figured that a different way of measuring receptive fields was necessary to test our theory.[10/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
But, we ran into some roadblocks. Many existing methods for measuring tuning curves rely on the STA. We show in our work that the STA is sensitive to biases arising from limited or incomplete sampling of the environment. [9/n]
31.07.2025 18:02 — 👍 1 🔁 0 💬 1 📌 0
While size heterogeneity in place fields has been documented, shape heterogeneity has been relatively understudied. So, we followed the predictions of our framework and tried to measure the degree of place field size and shape heterogeneity. [8/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
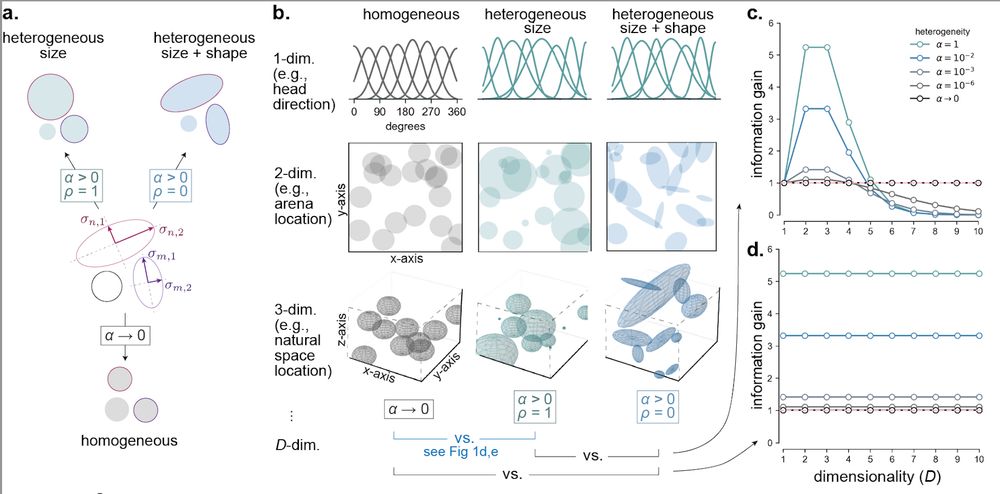
So, we introduced shape heterogeneity. When receptive fields vary in size and shape, populations can better encode 2- and 3-D quantities. But this property reverses for higher-dimensional stimuli. Size heterogeneity remains useful, while shape heterogeneity becomes harmful. [7/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
We dive into why this is in the manuscript, but I’ll spare the details here. This finding is fairly curious, because we know that place fields in rodents (encoding 2D spatial location) and bats (encoding 3D spatial location) have variably sized RFs… [6/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
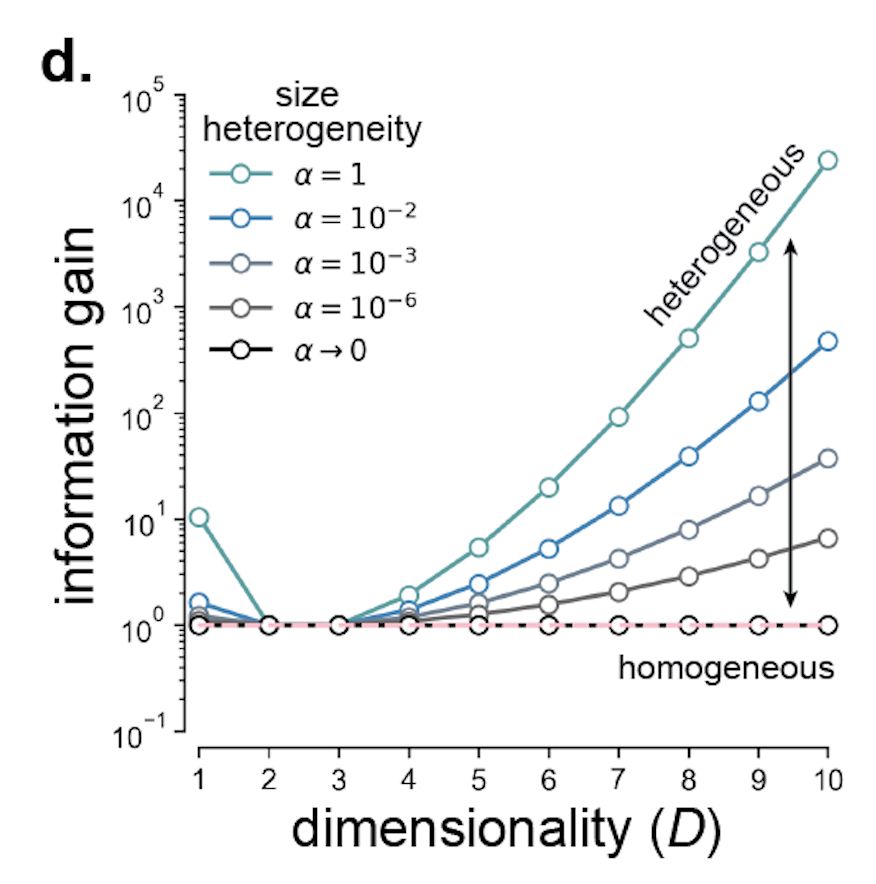
Let’s start with size heterogeneity, where we find dimensionality-dependence: Heterogeneously sized RFs increase info of one dimensional stimuli (head direction, eg). And >3-D stimuli. Strikingly, size heterogeneity isn’t helpful for encoding 2- or 3-D variables. [5/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
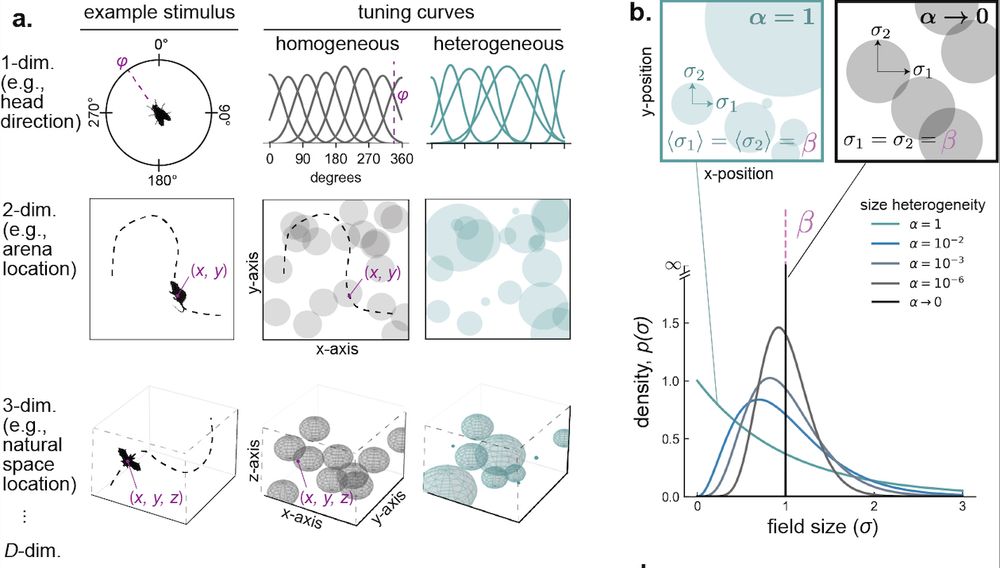
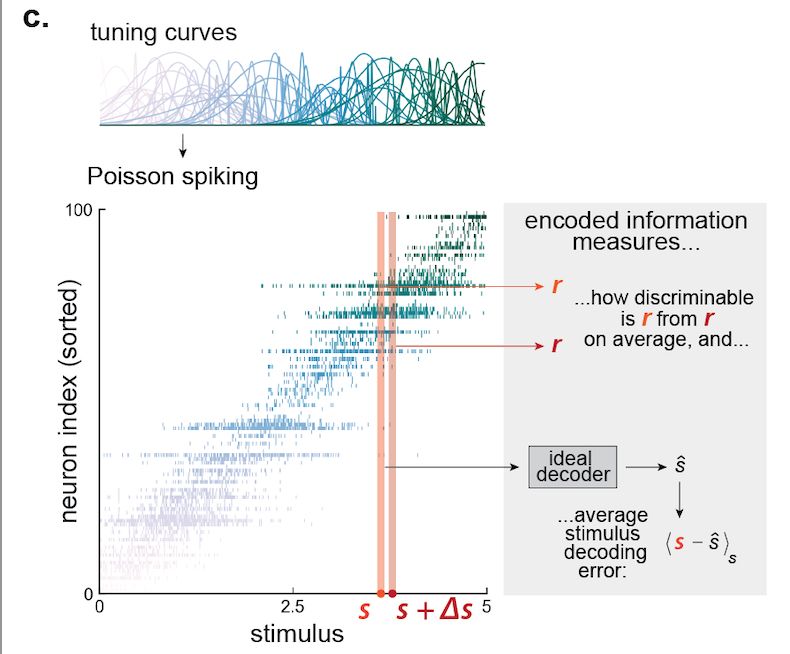
…until now! We derived the amount of information encoded about a stimulus in spiking activity as a function of the degree of the population’s RF heterogeneity. To make the theory general, we derived this relationship for arbitrary stimulus dimensionalities. [4/n]
31.07.2025 18:02 — 👍 1 🔁 0 💬 1 📌 0
Why would the brain prefer such variability? Is there a general coding benefit associated with heterogeneously sized receptive fields? This question has been asked in specific populations and under specific conditions, but a theory that unifies these accounts is lacking… [3/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
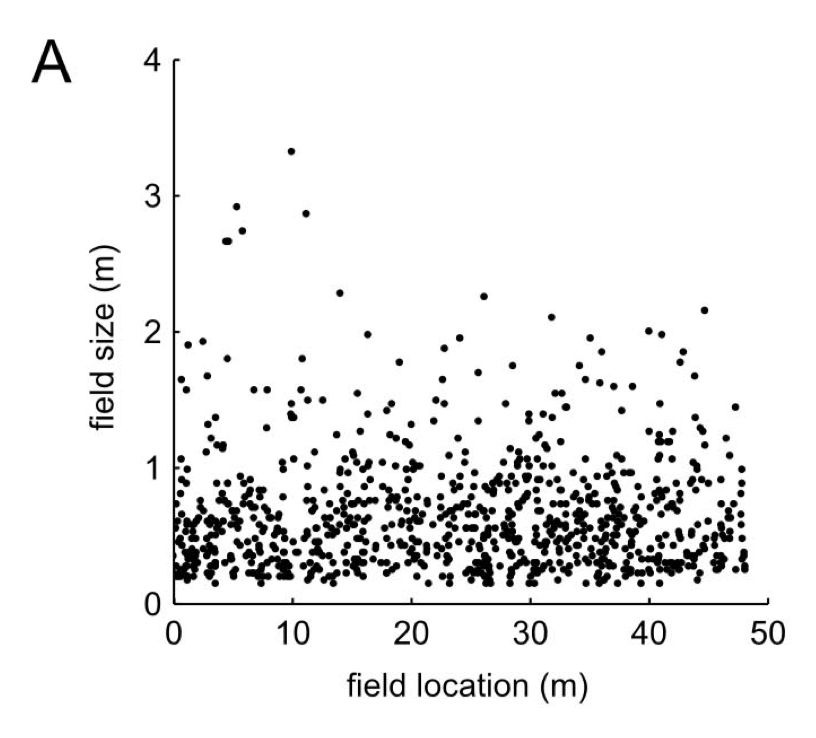
Populations across the brain have differently sized and shaped receptive fields. A striking example of this is hippocampal CA1 place cells, whose receptive fields can be mere centimeters across or as large as a few meters in size (fig from Rich et al., 2014) . [2/n]
31.07.2025 18:02 — 👍 0 🔁 0 💬 1 📌 0
Computational neuroscientist at WashU
https://www.hiratanilab.org/
Theoretical neuroscientist
Research fellow @ Kempner Institute, Harvard
dclark.io
Theoretical/Computational neuroscientist at Harvard Medical School | Algorithms and neural implementation of reasoning under uncertainty | Decision-making and navigation.
https://www.drugowitschlab.org/
HBI brings together neuroscience researchers from different parts of Harvard and its affiliated hospitals.
Throughout all that we do, we aspire to build and nurture a scientific community that is diverse, inclusive, and welcoming.
https://brain.harvard.edu
PhD Student at MIT Brain and Cognitive Sciences studying Computational Neuroscience / ML. Prev Yale Neuro/Stats, Meta Neuromotor Interfaces
Computational & Systems Neuroscience (COSYNE) Conference
🧠🧠🧠
Next: Mar 27-April 1 2025, Montreal/Tremblant
🧠🧠🧠
Here too:
@CosyneMeeting@neuromatch.social
@CosyneMeeting on Twitter
Biophysics PhD student with Jan Drugowitsch at Harvard Univ. (he/him) | theoretical neuroscience | spatial navigation under uncertainty
Postdoc @harvard.edu @kempnerinstitute.bsky.social
Homepage: http://satpreetsingh.github.io
Twitter: https://x.com/tweetsatpreet
PhD student @ Harvard || computational cognitive science, human decision making and reasoning
The Kempner Institute for the Study of Natural and Artificial Intelligence at Harvard University.
Computational neuroscientist interested in how we learn, and dad to twin boys
Asst prof at Baylor College of Medicine
https://www.henniglab.org/
Assistant Professor, McGill University | Associate Academic Member, Mila - Quebec AI Institute | Neuroscience and AI, learning and inference, dopamine and cognition
https://massetlab.org/
Senior Machine Learning Researcher, Kempner Institute for the Study of Natural and Artificial Intelligence, Harvard. Board Member, Neuromatch. she/her. Views are my own.
👩🏻🏫 asst teaching prof @ uc san diego
🧠 cognitive scientist
Theoretical neuroscientist; currently a Harvard Junior Fellow. jzv.io
Neuroscientist @Harvard asking how sensation and action are entwined to facilitate cognition. Natural behavior, circuits for scents and movement, curiosity, biological and artificial intelligence
Associate Professor at Harvard & Kempner Institute. Applying computational frameworks & machine learning to decode multi-scale neural processes. Marathoner. Rescue dog mom. https://www.rajanlab.com/
Neuroscientist
post-doc @zuckermanbrain.bsky.social @ Aronov lab 🪶
previously: phd @harvardmed.bsky.social with Nao Uchida
peruana & alfajor lover
News, commentary and research coverage from the international monthly journal publishing the highest quality of work in all areas of neuroscience.
https://www.nature.com/neuro/
Mood & Memory researcher with a computational bent. https://www.nicolecrust.com. Science advocate. Prof (UPenn Psych) - on leave as a Simons Pivot Fellow. Author: Elusive Cures. https://press.princeton.edu/books/hardcover/9780691243054/elusive-cures













