
Programmable antisense oligomers for phage functional genomics - Nature
Establishing antisense oligomers as versatile, non-genetic tools to silence phage mRNAs opens applications in basic research and biotechnology, as shown by identifying essential factors for propagatio...
No Genetics? Try ASOs – A non-genetic approach to silence genes at the phage-host interface. We use it to study jumbo phage biology and anti-phage defence.
@jorg-vogel-lab.bsky.social @helmholtz-hiri.bsky.social
@uni-wuerzburg.de @helmholtzhzi.bsky.social
published now in @nature.com
11.09.2025 09:10 — 👍 66 🔁 19 💬 0 📌 0
At last ! A tool specifically for phage-plasmid hunters. Check tyPPing by @karinailchenko.bsky.social and @eugenpfeifer.bsky.social
03.09.2025 14:00 — 👍 47 🔁 16 💬 0 📌 0
If you look for phage-plasmids, you absolutely have to try tyPPing!
Results and user guide on bioRxiv, GitHub+Zenodo
Check out: bsky.app/profile/kari...
Super proud (!) of the work by @karinailchenko.bsky.social & happy + many thanks to awesome collaboration with @epcrocha.bsky.social and R Bonnin.
04.09.2025 08:19 — 👍 5 🔁 0 💬 0 📌 0

Efficient detection and typing of phage-plasmids
Phage-plasmids are temperate phages that replicate as plasmids during lysogeny. Despite their high diversity, they carry genes similar to phages and plasmids. This leads to gene exchanges, and to the ...
🚨 New preprint! 🧬
𝐏𝐡𝐚𝐠𝐞-𝐩𝐥𝐚𝐬𝐦𝐢𝐝𝐬 (𝐏-𝐏𝐬) are fascinating elements: both 𝐭𝐞𝐦𝐩𝐞𝐫𝐚𝐭𝐞 𝐩𝐡𝐚𝐠𝐞𝐬 and 𝐩𝐥𝐚𝐬𝐦𝐢𝐝𝐬 ➡️ tricky to detect.
We present 𝐭𝐲𝐏𝐏𝐢𝐧𝐠 — the first 𝐢𝐝𝐞𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐚𝐧𝐝 𝐜𝐥𝐚𝐬𝐬𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐭𝐨𝐨𝐥 🛠️designed specifically for P-Ps:
✅ Accurate
✅ Sensitive
✅ Easy to use
📖 www.biorxiv.org/content/10.1...
03.09.2025 13:02 — 👍 31 🔁 17 💬 2 📌 2
Habemus paper! Our story on integron-encoded anti-phage defenses is now out in @science.org! 16 new systems, small versions of known ones, and a lot more in this highly-collaborative study.
Many thanks to everyone involved, especially my supervisor @epcrocha.bsky.social
bsky.app/profile/bapt...
09.05.2025 11:51 — 👍 33 🔁 18 💬 2 📌 1
Overall, our findings underscore the complex interplay within the gut microbiome, where antibiotics not only affect bacteria but also reshape the associated phage communities, and in turn again influence the bacterial populations.
We look forward to get further interesting insights in future works!
28.07.2025 08:41 — 👍 0 🔁 0 💬 0 📌 0
We believe that its phages are the key players!
By targeting these dominant bacteria, these phages create opportunities for other bacterial species to re-colonize, ultimately helping to regain diversity in the gut!
28.07.2025 08:41 — 👍 4 🔁 1 💬 1 📌 0
We believe that "Kill-the-Winner" dynamics are at play: Antibiotics create a disturbance allowing certain bacteria to flourish.
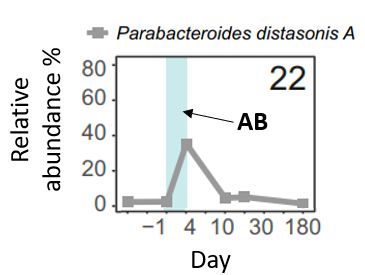
Parabacteroides distasonis is one of those, and typically thrives after cephalosporin treatment.
What wasn't fully understood was why it loses its dominance.
28.07.2025 08:41 — 👍 1 🔁 0 💬 1 📌 0

3rd: Despite the initial loss of some phage species, we saw a temporary and significant increase in the number of dominant, virulent phages. But why?
28.07.2025 08:41 — 👍 0 🔁 0 💬 1 📌 0

2nd: Gut phages are unique to each individual (nothing new) and here we show that your unique phage "fingerprint" largely persists even after strong perturbations. So, in a way, your phages remain truly yours!
28.07.2025 08:41 — 👍 0 🔁 0 💬 1 📌 0

We studied the dynamics of gut phage populations in healthy individuals who received antibiotic treatment: 3rd Gen Cephalosporins.
1st: Antibiotics💊do not only just affect bacteria🦠 but also their viruses. We observed a 20% decrease in the richness+diversity, but (lucky us) it recovers over time!
28.07.2025 08:41 — 👍 0 🔁 0 💬 1 📌 0
Excited 🥳 to share our latest work on gut phages!
Big thanks to @epcrocha.bsky.social, Erick D, Camille d'H, @fplazaonate.bsky.social, Quentin LB, and all others involved for support and contributions! 🙌
Out in Cell Reports @cp-cellreports.bsky.social
doi.org/10.1016/j.ce...
Here's what we found 🤓
28.07.2025 08:41 — 👍 31 🔁 27 💬 1 📌 1
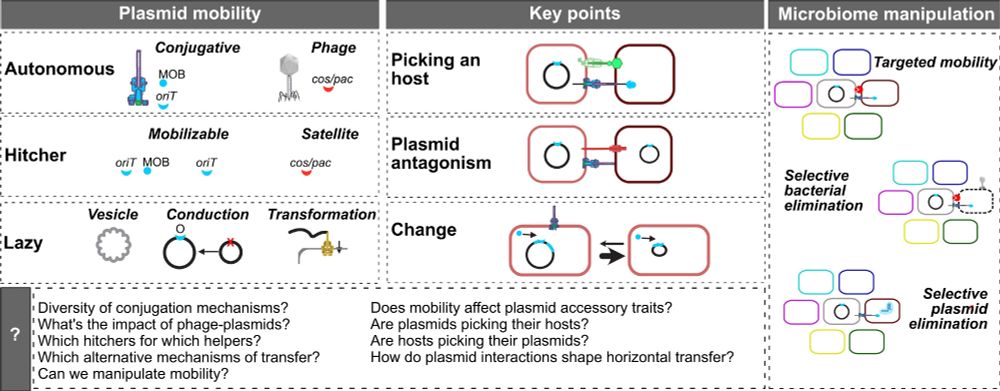
graphical abstract of the article the extended mobility of plasmids
Here's our new broad review on the extended mobility of plasmids, about all mechanisms driving and limiting their transfer. From conjugation to conduction, phage-plasmids to hitchers, molecular to evolutionary dynamics, ecology to biotech. The state of affairs. 1/9 academic.oup.com/nar/article/...
23.07.2025 07:35 — 👍 184 🔁 93 💬 4 📌 9

GitHub - EpfeiferNutri/PrediRes
Contribute to EpfeiferNutri/PrediRes development by creating an account on GitHub.
This work sheds light on the underappreciated role of phages in post-antibiotic gut microbiome recovery. We think this is just the beginning!
More infos here: github.com/EpfeiferNutr...
and👇
doi.org/10.5281/zeno...
10.02.2025 09:55 — 👍 2 🔁 0 💬 0 📌 0

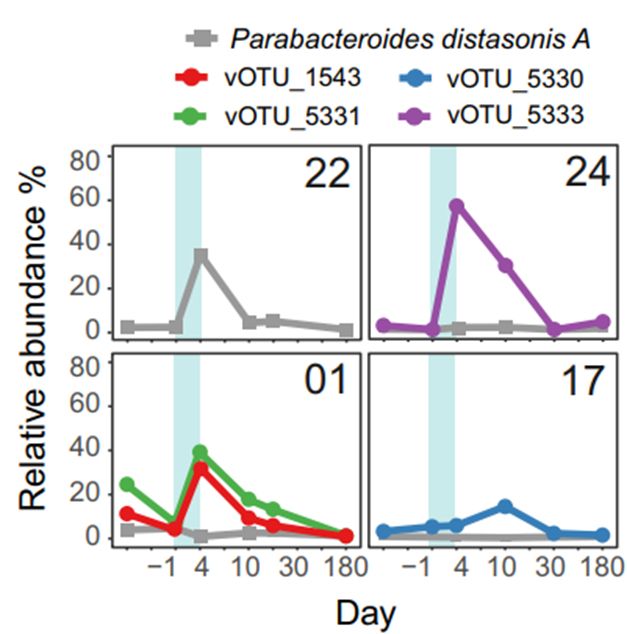
A great example:
Parabacteroides distasonis known to bloom after treatment🦠➡️🔋and only when its phages were absent.
But when they (phages) were around and burst 📈 P. distasonis bacteria were hardly detectable 🦠➡️🪫
10.02.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0

🧐We believe these phages help gut recovery! 🛡️
They prevent bacterial blooms of (e.g., antibiotic-resistant) species that would otherwise dominate the gut.
10.02.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0

💥 Most surprising: some phages thrived the day after treatment! More than on any other day.
🚀 BUT: these phages were virulentand NOT induced prophages. So, what is their role 🤔?
10.02.2025 09:55 — 👍 2 🔁 1 💬 1 📌 0

So, what happens to gut phages after antibiotics?🤔 20% of them vanished after treatment 📉, but over time, recovery occurred
📌The response was highly individual-specific, reinforcing the uniqueness of each person’s microbio- and phageome!
10.02.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0

We found >6400 phage species🧮, lots of high quality🌟, most predicted to be temperate, ca. 1900🆕 ones, and (super exciting!) a lot are also phage-plasmids!
10.02.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0

🧐We fine-tuned the analysis of the phageome part taken from the CEREMI trial🔍
Briefly: 22 volunteers (healthy background) received antibiotic treatment💊 typical for a clinical setting. Their 💩 were analyzed over a period up to 180 days after treatment for phages and bacteria
10.02.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0
New preprint. Large genomes vibriophages have it all: multiple functions, autonomy (full set of tRNA genes!), anti-defenses, broad host range. Yet, they remain rare, pinpointing the limits of generalists. Great collaboration with @fredoleroux.bsky.social led by Charles Bernard in our lab.
01.01.2025 15:07 — 👍 43 🔁 27 💬 3 📌 1
Postdoc | bacterial defence systems ⚔️🛡️🦠
I've worked on all of science, from T cells to B cells.
https://fellowsherpa.com
Professor at the University of South Florida. Marine microbiology and virology, with a smattering of eukaryotes here and there because focus is for losers. Characterizing the global virome, one virus at a time. Owned by a very cute dog and cat. She/her
Bluesky shadowbanned me
Follow my alt account
Current yeast articles everyday :)
Follow our Feed #yeast #saccharomyces
:)
From Molecular Genetics Lab, USACh
PhD in Biology (Génétique et Génomique)
Evolution of integrons, in the lab of @epcrocha.bsky.social, Microbial evolutionary genomics @pasteur.fr
@EPFL and @Polytechnique alumni
Phage Group @ Laboratoire de Chimie Bactérienne
#phage genomics, #phage-host interactions, #phage-host coevolution
Microbiology Masters 🦠🔬 member of gator nation 🐊
🎾👟🚲
Left-handed & weekend cyclist. Assoc. Prof. Biochemistry & Bioinformatics @uammadrid.bsky.social and PI @RnR_Lab
DNA replication & repair: mechanisms, evolution and biotech 🚲💻🧬🧪
https://linktr.ee/mredrejo
Researching the diversity and interaction of phages with plant pathogenic bacteria 🌱🔬 | INRAE | PHIM | Montpellier, France. 🇵🇸
Post-doc - mosquito behavior @RadboudUMC in Nijmegen with Felix Hol. Previously IVIst with Louis Lambrechts lab @Institut Pasteur. Decoding bugs and/or debugging code.
Bacterial Genomes Adaptation_ CBI-Université de Toulouse 🇨🇵
🧬🦠 Postdoc AMR/Microbial Genomics/Bioinformatics. Tracking AMR bacteria and its MGEs from a One-Health perspective. Passionate ➡️ phylogenomics, GDL, TDL, deep learning, graphs, plasmids, phages,... 🇦🇷 in 🇨🇭
Population genetics, monke and life
Postdoc at U of Cambridge MRC Tox🇬🇧 PhD UWA🇦🇺Biotech UNQ🇦🇷🧬🛠️ Functional genomics + engineering biology to explore how microbes interact and unlock microbiome potential. She/her Latinx https://scholar.google.com/citations?user=RNoSH60AAAAJ&hl=en
PhD student studying Infant gut phages | Vatanen & Friman labs University of Helsinki | Viral dark matter | phage-host interactions | MGE competition | microbial ecology
Microbiology Society: A world in which the science of #microbiology provides maximum benefit to society | microbiologysociety.org
lnk.bio/microbiosoc


















