
Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
@minimolecule.bsky.social
Senior Postdoc at Dawson Lab, Peter Mac & Hon. Senior Research Fellow at UCL Cancer Institute | non-genetic resistance, gene regulation, and non-coding mutations in cancer bit.ly/3B0wvaA - Google Scholar Narrm (Melbourne)/London

Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
Congrats Nicola, brilliant work!
18.04.2025 10:53 — 👍 0 🔁 0 💬 0 📌 0
Starting off my bluesky journey by sharing my paper published this week: CD21 targeted CAR-T cells for the treatment of T-ALL.
The result of many years of work from many people. Funded by #CRUK #UKRI MRC and #GOSHCC
www.science.org/doi/10.1126/...
My previous mentor and great friend Prof. Marc Mansour from UCL has joined Bluesky! 🎉 Do consider following him if you’re researching acute leukaemias and associated novel therapies!
bsky.app/profile/marc...
Check out our new pre-print!
Exciting times ahead for Menin biology and targeting epigenetics regulators in AML.
Congratulations to all involved!
Thanks Marina, too kind. Such fond memories of those committee meetings...those were the days... 💙
10.12.2024 10:57 — 👍 0 🔁 0 💬 0 📌 0This research was funded by Leukaemia UK, @cancerresearchuk.org, GOSH Charity and others. None of these discoveries would be possible without the support and consent from patients and their families. This discovery is for them.
10.12.2024 10:34 — 👍 1 🔁 0 💬 0 📌 0A huge thanks to all collaborators involved for their expert input, most haven't come over to 🦋 as yet bar the wonderful @adelekfielding.bsky.social
10.12.2024 10:34 — 👍 1 🔁 0 💬 2 📌 0I want to highlight the incredible support of
@mafdawson.bsky.social who has welcomed me into his lab at a critical point in my academic career and my previous mentor Prof. Marc Mansour (UCL) for his advice and guidance on this project and beyond.
A special thanks to the incredibly talented Gianna Bloye who completed critical parts of this paper and is now doing her PhD @ox.ac.uk. And to Nadine Farah (UCL) and Jonas Demeulemeester (VIB-KU) who have been on this challenging expedition with me to find cis-acting noncoding mutations in T-ALL.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0We speculate that ‘promoter tethering’ of oncogenes to inert regions of the genome is a previously unappreciated mechanism preventing tumorigenesis, and postulate similar mechanisms may be found in other cancers.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0These findings also add to the complex regulatory relationship between the FTO and IRX3 genes, first identified through the discovery of obesity-associated germline variants.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
This mechanism differs to previously described enhancer hijack events, which bring enhancers and promoters together through structural rearrangement. And differs to examples of focal deletions that impinge on TAD boundaries as we believe this all happens intra-TAD.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0These data suggest IRX3 is sequestered to FTO intron 8 through a ‘promoter tether’ facilitated by CTCF. Loss of this CTCF site by focal deletion untethers the IRX3 promoter allowing for enhancer hijack. This may be an example of E-P competition occurring within the same TAD.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
By using CRISPR/Cas9 we disrupted the CRNDE super-enhancer in FTO intron 8 CTCF site deleted cells (ALLSIL). This led to a significant reduction in IRX3 expression, meaning this super-enhancer is indeed hijacked by IRX3.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
So what’s the deal with CRNDE? It encodes for a lncRNA that is actively transcribed throughout T cell development and harbours a super-enhancer. HiChIP shows this super-enhancer loops to IRX3 suggesting its hijacked in FTO intron 8 CTCF site deleted T-ALL cells (ALLSIL)
10.12.2024 10:34 — 👍 1 🔁 0 💬 1 📌 0
We also baited the IRX3 promoter in our CRISPR/Cas9 edited cells (PF382) with and without the FTO intron 8 CTCF site, observing the same effect of increased contacts with the CRNDE region along with increased transcriptional output of IRX3.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0In contrast doing the same with FTO intron 8 CTCF site deleted cells (ALLSIL) massively increased the contacts between the IRX3 promoter and CRNDE along with transcriptional activation of IRX3. Look at the contact delta 👆(above)
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
With UMI-4C we baited the CTCF site in FTO wild-type cells (PF382). This showed that IRX3 is ‘tethered’ to FTO intron 8. In the same cells we baited the IRX3 promoter which identified minimal contacts with CRNDE with no transcriptional output of IRX3.
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0So what does the FTO intron 8 CTCF site usually do? 🤔
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
CRISPR/Cas9-mediated deletion of the FTO intron 8 CTCF site in IRX3 negative (PF382) T-ALL cells transcriptionally activated IRX3 in single cell sorted clones and polyclonal populations, with no activation if you only delete the MYB site. This suggested the CTCF site was critical.
10.12.2024 10:34 — 👍 1 🔁 0 💬 1 📌 0Whilst most patients had copy number loss for both CTCF and MYB sites, 4/12 had heterozygous copy number loss of the CTCF site alone, suggesting that loss of this CTCF site was more likely to be functionally relevant. But do the deletions transcriptionally activate IRX3?
10.12.2024 10:34 — 👍 1 🔁 0 💬 1 📌 0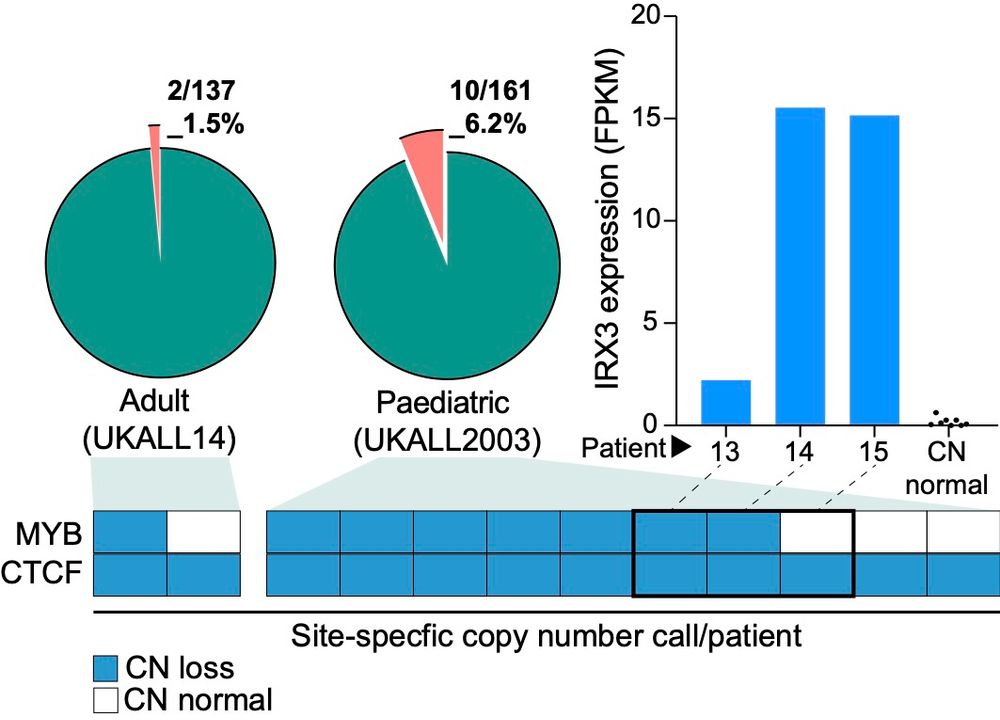
By using ddPCR probes against the CTCF and MYB sites we identified CN deletions in a site-specific manner. This showed FTO intron 8 deletions occur in 1.5% of adults and 6.2% of paediatric patients with T-ALL. But have a look at the site-specific calls👀
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
Amazingly these deletions defined a minimal region of ~155kb. ChIP-seq data from a T-ALL cell line identified CTCF binding and a MYB bound enhancer within this region. Both CTCF and MYB have previously been implicated in mechanisms of aberrant gene expression in T-ALL
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
Expression data was available for 6/13 T-ALL samples showing these were IRX3+ by RNAseq. Notably, 1 patient exhibited allele specific expression supporting our working hypothesis that IRX3 expression may be driven by a cis-acting genetic lesion
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
By studying published genomic datasets from primary T-ALL patient samples and T-ALL cell lines we identified 13 T-ALL genomes (12 from patients and 1 cell line) with heterozygous copy number losses impinging on FTO intron 8 – the 3’ contact point to IRX3
10.12.2024 10:34 — 👍 1 🔁 0 💬 1 📌 0
IRX3 sits in a single TAD with neighbouring genes FTO, CRNDE and IRX5. HiChIP data from an IRX3 positive cell line identified contacts 3’ to IRX3 within FTO intron 8, and 5’ to IRX3 within the CRNDE/IRX5 locus. We thought these two contact points may hold the clue…
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0
IRX3 is not expressed in normal T-cells. But high IRX3 expression is a feature of T-ALL and observed across differing molecular subtypes of T-ALL. Special mention to Somervaille Lab @cruk-mi.bsky.social who have previously shown it can cause T-ALL in vivo. The question is what’s driving this?
10.12.2024 10:34 — 👍 1 🔁 1 💬 1 📌 0
Link here to the manuscript if you want to get right into it, or carry on to the thread below. If you're into gene-regulation and/or noncoding mutations this may be of particular interest! (Note: I'm bringing this thread over from the other place).
10.12.2024 10:34 — 👍 0 🔁 0 💬 1 📌 0For my new followers! Focal deletions of noncoding regions in cancer genomes can have unexpected consequences. Out now in @bloodjournal.bsky.social, we’ve discovered a novel mechanism of oncogene activation whereby focal deletion of a ‘promoter tether’ leads to aberrant expression of IRX3 in T-ALL.👇
10.12.2024 10:34 — 👍 16 🔁 5 💬 2 📌 2