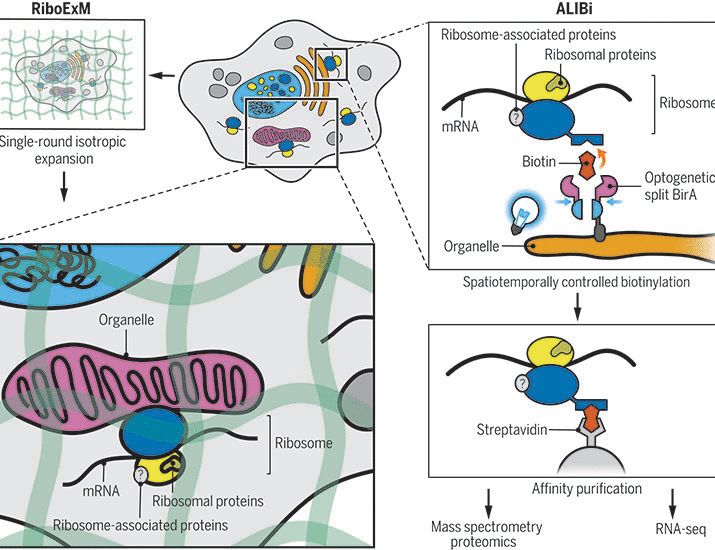
The lab of Single cell ecology has just opened its doors at the University of Glasgow, School of Biodiversity, One Health and Veterinary Medicine!
Join us, collaborate or learn about what we do:
mancinilab.github.io/index.html
And check out our most recent output:
www.biorxiv.org/content/10.1...
02.12.2025 10:33 — 👍 2 🔁 1 💬 0 📌 1

It was our pleasure to host @james-bryson.bsky.social (BRIC, Copenhagen) at @crg.eu through the EU-LIFE Postdoctoral Exchange Program!
Big thanks to James for presenting his work on advanced CRISPR tools for epitranscriptomic research, and to @eu-life.bsky.social for this opportunity!🧬✈️
#EULIFE #CRG
10.11.2025 15:59 — 👍 9 🔁 5 💬 0 📌 0
@embl.org is undoubtedly one of the best place in the world to start your lab! The GB unit is collegial, supportive, collaborative and the science is just outstanding. Apply! DM me if you have questions.
30.07.2025 14:01 — 👍 14 🔁 7 💬 1 📌 0
Sciencing in the summer
11.07.2025 16:44 — 👍 1 🔁 0 💬 0 📌 0
Would you pay $150 per article to post your preprint on @bioRxiv @aRxiv @medRxiv @chemRxiv etc.? Currently it’s free but a small cost could help offset running costs and also work as a barrier against paper mills and AIs just posting all the time and filling these sites up.
09.07.2025 18:03 — 👍 88 🔁 19 💬 25 📌 5
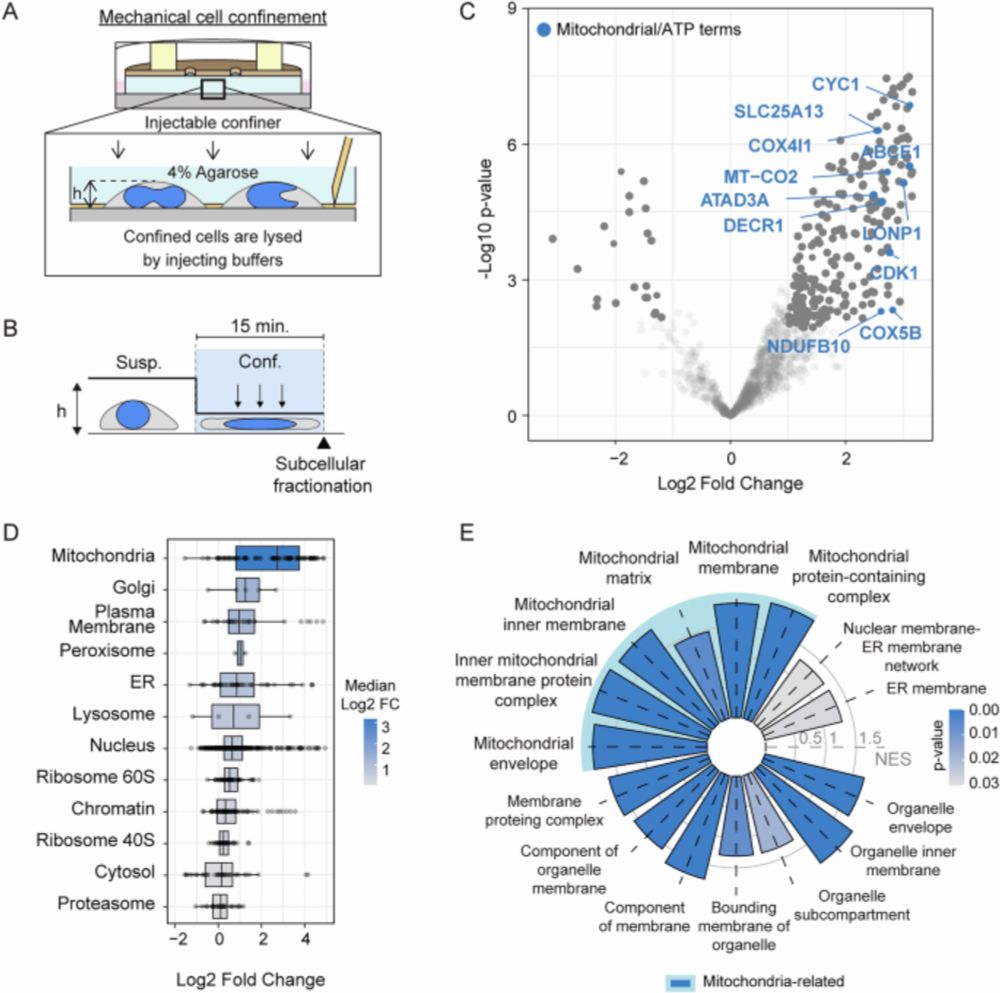
Decoding cnidarian cell type gene regulation
Animal cell types are defined by differential access to genomic information, a process orchestrated by the combinatorial activity of transcription factors that bind to cis -regulatory elements (CREs) to control gene expression. However, the regulatory logic and specific gene networks that define cell identities remain poorly resolved across the animal tree of life. As early-branching metazoans, cnidarians can offer insights into the early evolution of cell type-specific genome regulation. Here, we profiled chromatin accessibility in 60,000 cells from whole adults and gastrula-stage embryos of the sea anemone Nematostella vectensis. We identified 112,728 CREs and quantified their activity across cell types, revealing pervasive combinatorial enhancer usage and distinct promoter architectures. To decode the underlying regulatory grammar, we trained sequence-based models predicting CRE accessibility and used these models to infer ontogenetic relationships among cell types. By integrating sequence motifs, transcription factor expression, and CRE accessibility, we systematically reconstructed the gene regulatory networks that define cnidarian cell types. Our results reveal the regulatory complexity underlying cell differentiation in a morphologically simple animal and highlight conserved principles in animal gene regulation. This work provides a foundation for comparative regulatory genomics to understand the evolutionary emergence of animal cell type diversity. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, https://ror.org/0472cxd90, ERC-StG 851647 Ministerio de Ciencia e Innovación, https://ror.org/05r0vyz12, PID2021-124757NB-I00, FPI Severo Ochoa PhD fellowship European Union, https://ror.org/019w4f821, Marie Skłodowska-Curie INTREPiD co-fund agreement 75442, Marie Skłodowska-Curie grant agreement 101031767
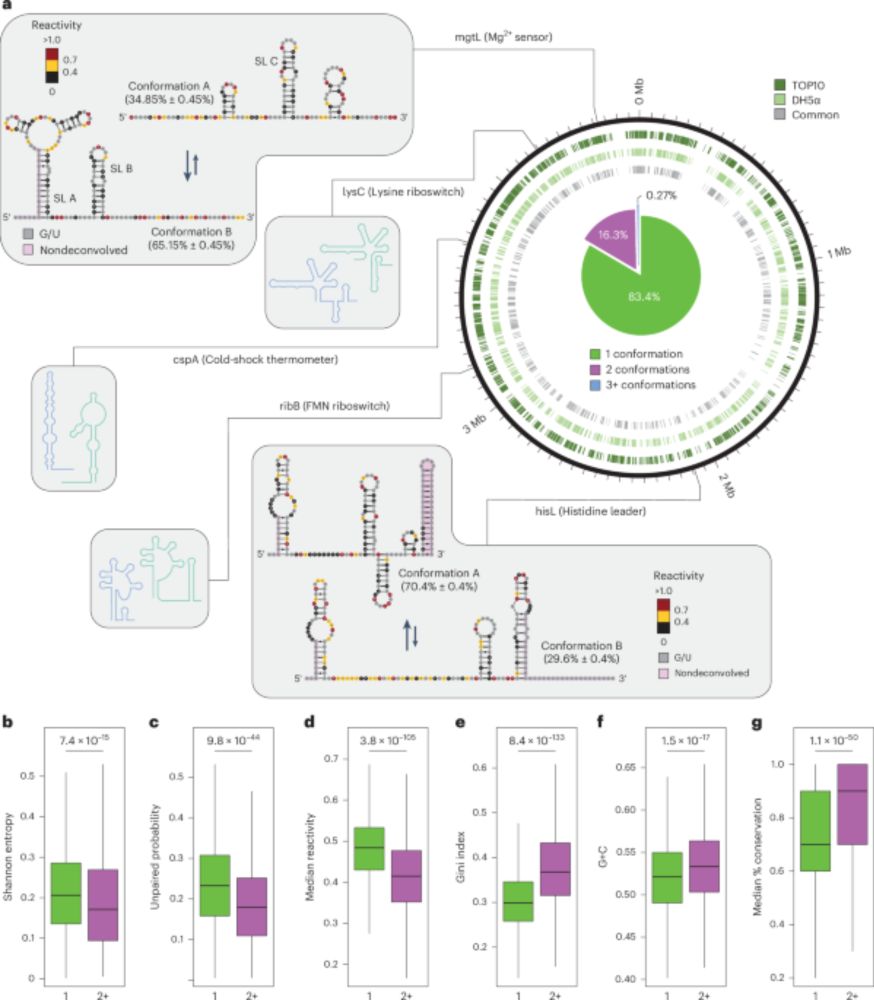
I am very happy to have posted my first bioRxiv preprint. A long time in the making - and still adding a few final touches to it - but we're excited to finally have it out there in the wild:
www.biorxiv.org/content/10.1...
Read below for a few highlights...
06.07.2025 18:14 — 👍 59 🔁 24 💬 1 📌 2

Endinsa’t en una vivència sensorial única que connecta ciència i art immersiu. “SEEING: Un ull i una caixa” arriba del 5 al 14 de juny al Centre Cívic Convent de Sant Agustí (Carrer del Comerç 36, Barcelona). www.crg.eu/en/event/see...
30.05.2025 11:24 — 👍 9 🔁 17 💬 0 📌 1
Our genome encodes > 7000 microproteins that have been previously ignored in genome annotations. How do these microproteins impact tissue function? Here, we use an in vivo single-cell CRISPR screen to systematically explore microprotein function in the mouse epidermis. 1/9
19.03.2025 15:44 — 👍 67 🔁 17 💬 3 📌 5
Scientific research is a strategic necessity for resilience and competitiveness in Europe!
"While the minimum basis for a thriving European Research & Innovation is a stand-alone, well budgeted #FP10, we urge the European Commission to further follow 5 key actions"
lnkd.in/d2mzN_zu
19.03.2025 10:22 — 👍 5 🔁 4 💬 0 📌 0
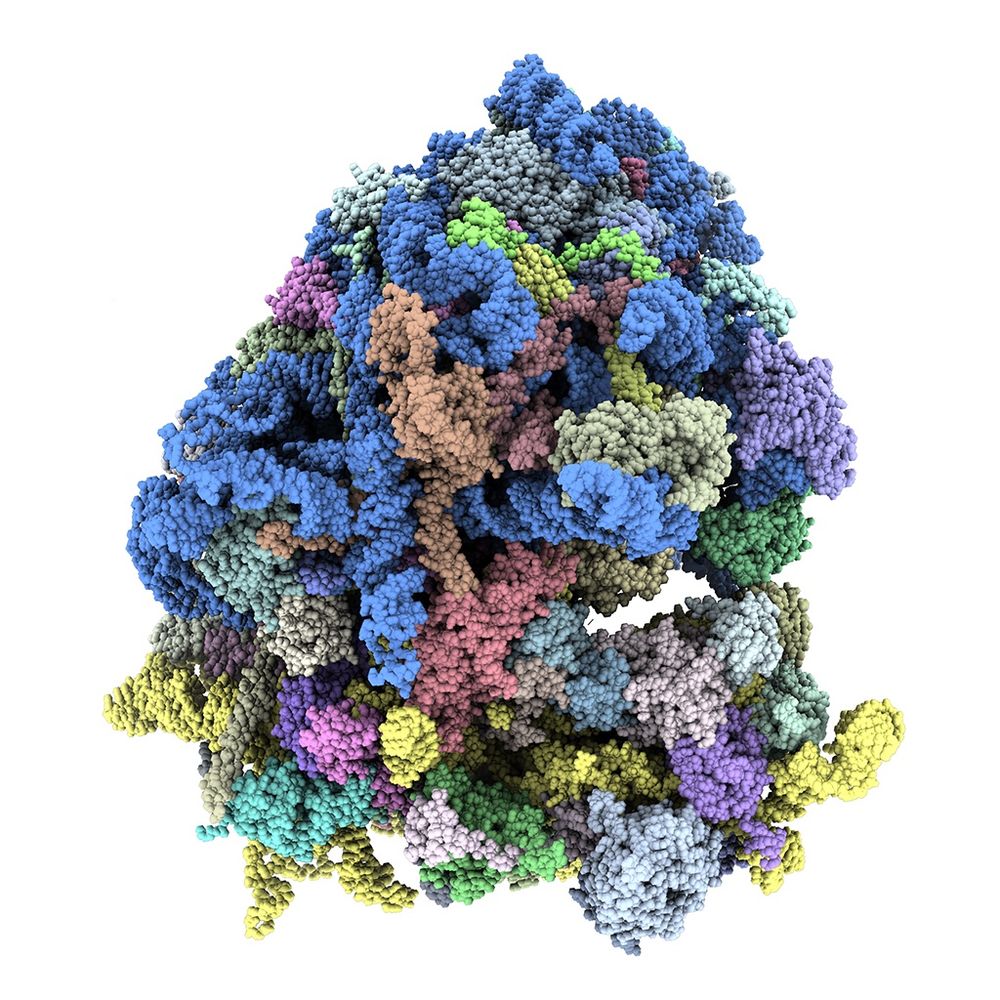
Philosophical Transactions of the Royal Society B: Biological Sciences: Vol 380, No 1921
Excited to see issue on ribosome heterogeneity and specialisation out @royalsociety.org Philosophical Transactions B. Thanks to authors & co-editors @liamfaller.bsky.social @andershlund.bsky.social Maria Barna. Wonderful cover Bulat Fatkhullin! #ribosomes royalsocietypublishing.org/toc/rstb/202...
06.03.2025 14:59 — 👍 14 🔁 10 💬 0 📌 2

New manuscript from the Novoa lab was published this week in Genome Biology! You can give it a read here: rdcu.be/ebiIs
28.02.2025 16:18 — 👍 14 🔁 7 💬 2 📌 0
Great comparison of the experience @crg.eu vs Babraham Institute by Hanane! And congrats to Shota (@trayon.bsky.social lab) and Alex (@houseleylab.bsky.social) for their poster and presentation successes! #CRGBIpostdocs
03.03.2025 09:40 — 👍 3 🔁 1 💬 0 📌 0
METIS is a research group in analytic philosophy based at the Department of Logic, History, and Philosophy of Science of UNED, in Madrid. Find out more about us at blogs.uned.es/metis
Posts by: @ignasigil.bsky.social & @raciondeser.bsky.social
*Views are my own and possibly wrong
📖Senior Editor at BMC Biology
🦠Part-time community college prof
🧘🏻♀️200 hour RYT
📍 Philadelphia
Past:
🔬Postdoc Thomas Jefferson University
🧬PhD @stonybrooku.bsky.social
🧪BS @mnstatemankato.bsky.social
Cancer Research UK Scotland Institute (formerly the CRUK Beatson Institute). Our Mission: Cancer discovery for patient benefit.
www.crukscotlandinstitute.ac.uk
Group leader at EMBL Heideberg
https://www.embl.org/groups/krebs/
Also known as the ribosome company, we are dedicated to advancing knowledge of RNA and ribosome biology by enabling scientists to study the mechanisms of translation in action.
https://www.immaginabiotech.com
The Coalition for Advancing Research Assessment (CoARA) is a collective of organisations committed to reforming research evaluation practices in line with the Agreement (ARRA). Learn more at www.coara.eu and join the coalition!
European Forum for Studies of Policies for Research and Innovation:
interdisciplinary research on policy and governance around knowledge creation and innovation
professor @dartmouth + science policy @ifp //
leveraging research to promote science and progress
Co-founder IFP // scientific, technological, and industrial progress
Living my purpose
Writer:
Books #LessonsFromPlants (https://www.hup.harvard.edu/books/9780674241282) & #WhenTreesTestify (https://us.macmillan.com/books/9781250335166/whentreestestify/)
Professor
#Leadership|#Mentoring|#EquityReadingList/#DEIReadingList
Author of SWIFTYNOMICS: How Women Mastermind and Redefine Our Economy 📚 Order here: https://a.co/d/dDydwxe. Currently: co-director Kansas Pop Center & associate professor, KU. Formerly: Census|NIH|USDOL.
undisciplined scholar and recovering academic @AEI @DNVA1 @CUBoulder @UCL | Subscribe to The Honest Broker ➡️ http://rogerpielkejr.substack.com
VP/CSO @HHMI.bsky.social & Head of #VosshallLab @rockefelleruniv Toward Excellent, Inclusive, & Open Science
Science communicator: Passionate about science, family, endless forms most beautiful...and Swiss chocolate. Author of WOMEN IN SCIENCE NOW (https://amazon.com/Women-Science-Now-Strategies-Achieving/dp/0231206143) via
@columbiaup.bsky.social
Executive Director, Good Science Project
West Coast Biotech & Life Sciences Reporter for Stat News. Stanford Immunology PhD & UCSC SciCom alum. Tips? jonathan.wosen@statnews.com. Signal: jwosen.27
Professor and Chair of WashU Chemistry. I’m a researcher, but I’m a #MentorFirst. Author of Labwork to Leadership.
Open Science Advocate | Senior Program Lead, MIT Open Learning
Somerville, MA
Former Harvard Fellow, former chief @SpeakerPelosi energy climate science and tech advisor