
ChemEmbed: a deep learning framework for metabolite identification using enhanced MS/MS data and multidimensional molecular embeddings url: academic.oup.com/bib/article/...
13.02.2026 21:59 — 👍 4 🔁 4 💬 0 📌 0@seeslab.bsky.social
Marta Sales-Pardo & Roger Guimerà. Complex systems & networks; Statiscal learning; Comput. social science; Systems biology at @universitatURV @icreacommunity

ChemEmbed: a deep learning framework for metabolite identification using enhanced MS/MS data and multidimensional molecular embeddings url: academic.oup.com/bib/article/...
13.02.2026 21:59 — 👍 4 🔁 4 💬 0 📌 0
🚀A ICREA tornem a estar actius a les xarxes socials! Parlarem de:
✨ Nous descobriments de la #ComunitatICREA
📢 Convocatòries
🎉 Esdeveniments i congressos
🧪 I molta recerca d'excel·lència!
Segueix-nos a X x.com/icreacommunity i LinkedIn www.linkedin.com/company/icrea i no et perdis cap novetat!
🧵
1/ New from @reeserichardson.bsky.social, @jabyrnesci.bsky.social, and our lab in @pnas.org :
A growing body of evidence shows that "systematic" scientific fraud is an emerging threat to the integrity of science.
Our latest study investigates how this fraud is organized and sustained.

New paper out in Briefings in Bioinformatics
📰SingleFrag: a deep learning tool for MS/MS fragment and spectral prediction and metabolite annotation academic.oup.com/bib/article/...
Yet another great collaboration with @oyanes.bsky.social
14.07.2025 14:54 — 👍 1 🔁 0 💬 0 📌 0
As a proof of concept, we successfully annotate three previously unidentified compounds frequently found in human samples
14.07.2025 14:54 — 👍 0 🔁 0 💬 1 📌 0
Our results demonstrate that SingleFrag surpasses state-of-the-art in silico fragmentation tools, providing a powerful method for annotating unknown MS/MS spectra of known compounds
14.07.2025 14:54 — 👍 0 🔁 0 💬 1 📌 0Here, we present #SingleFrag, a novel deep learning tool that predicts individual MS/MS fragments separately, rather than attempting to predict the entire fragmentation spectrum at once
14.07.2025 14:54 — 👍 1 🔁 0 💬 1 📌 0Identifying metabolites in MS/MS data often means matching experimental spectra with existing spectral libraries. But, with limited libraries, identifying unknowns remains a big hurdle in #metabolomics
14.07.2025 14:54 — 👍 0 🔁 0 💬 1 📌 0
New paper out in Briefings in Bioinformatics
📰SingleFrag: a deep learning tool for MS/MS fragment and spectral prediction and metabolite annotation academic.oup.com/bib/article/...

SingleFrag: a deep learning tool for MS/MS fragment and spectral prediction and metabolite annotation #BriedBioinform academic.oup.com/bib/article/...
11.07.2025 18:39 — 👍 8 🔁 2 💬 0 📌 0Thanks 🙏🙏 @martikagv.bsky.social @vcolizza.bsky.social @pessoabrain.bsky.social @asteixeira.bsky.social @gomezgardenes.bsky.social @seeslab.bsky.social (and all the speakers not in this platform) for their amazing contribution to make this edition a reference for young Network Scientists.
05.07.2025 10:11 — 👍 6 🔁 1 💬 0 📌 0
@mscxnetworks.bsky.social ended.
A week of cutting edge complexity science, from foundations to applications. Amazing speakers and great cohort of attendants.
One of the best editions ever. 10 years of passion, love and network science.
In an amazing piece of #Sicily
youtu.be/Nh5vrEKheH0?...
Manuel Ruiz-Botella, Marta Sales-Pardo, Roger Guimer\`a: A collaborative constrained graph diffusion model for the generation of realistic synthetic molecules https://arxiv.org/abs/2505.16365 https://arxiv.org/pdf/2505.16365 https://arxiv.org/html/2505.16365
23.05.2025 06:06 — 👍 1 🔁 4 💬 1 📌 0Great work by first author Manuel Ruiz-Botella!
25.06.2025 14:15 — 👍 0 🔁 0 💬 0 📌 0
Leveraging the model’s efficiency, we created a database of 8.2M synthetically generated molecules and conducted a Turing-like test with organic chemistry experts to further assess the plausibility of the generated molecules, and potential biases and limitations of #CoCoGraph
25.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

#CoCoGraph outperforms state-of-the-art approaches on standard benchmarks while requiring up to an order of magnitude fewer parameters
25.06.2025 14:15 — 👍 1 🔁 0 💬 1 📌 0
New preprint out in the arXiv!
We introduce #CoCoGraph, a collaborative and constrained graph diffusion model capable of generating molecules that are guaranteed to be chemically valid www.arxiv.org/abs/2505.16365

💡Our paper on probabilistic alignment of networks has been highlighted by Nature Communications as one the 50 best papers recently published in the area of Applied physics and mathematics www.nature.com/collections/...
📰Read the paper www.nature.com/articles/s41...

At #NetSci2025 @netsciconf.bsky.social today? Don't miss Gemma Bel's poster at the Network Neuroscience satellite
📰Model-based alignment of developing connectomes
📍FaSos GG76S 1.018
🕔5:30pm

At #NetSci2025? Don't miss Teresa Lazaros's talk today at the Network Neuroscience satellite
📰 Probabilistic network alignment applied to brain connectomes
📍 FaSos GG76S 1.018
🕔 5pm
🔗 to paper: dx.doi.org/10.1038/s414...

At #NetSci2025 today? Don't miss Manuel Ruiz-Botella's talk at the @netbiomed2025.bsky.social satellite
📰 CoCoGraph: A collaborative constrained graph diffusion model for the generation of realistic synthetic molecules
📍 FaSos FaSoS GG76 1.02
🕒 3:30pm
🔗 to paper: doi.org/10.48550/arX...

#Networks www.nature.com/articles/s41...
16.05.2025 15:52 — 👍 1 🔁 1 💬 0 📌 0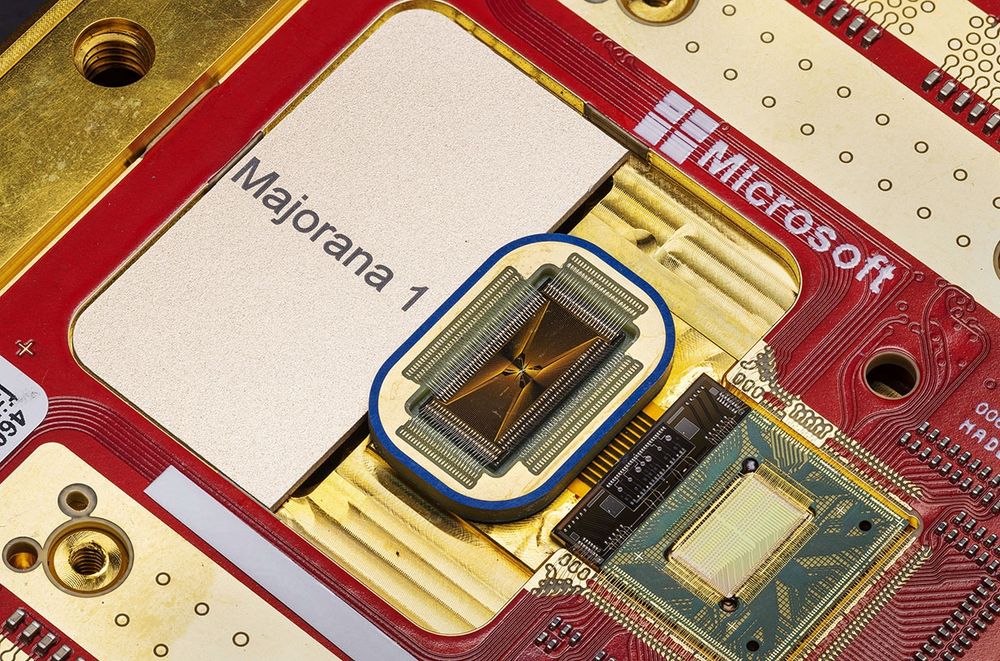
‘Data manipulations’ alleged in study that paved the way for Microsoft’s quantum chip | Science | AAAS www.science.org/content/arti...
12.05.2025 05:16 — 👍 1 🔁 0 💬 0 📌 0
So happy to share this one! Beautiful collaboration with @anduviera.bsky.social, @raissadsouza.bsky.social and Guram Mikaberidze on network flows: journals.aps.org/prx/abstract...
08.05.2025 21:08 — 👍 6 🔁 5 💬 0 📌 1We're looking for a new colleague to join us in Maastricht as an Assistant/Associate Professor in statistical learning in our Data Analytics and Digitalisation department.
Deadline to apply: June 8th
vacancies.maastrichtuniversity.nl/job/Maastric...

New substack post: I discuss a recent innovative method—ProbAlign, introduced by Lázaro, Guimerà, & Marta Sales-Pardo—which uses probabilistic modeling to achieve more accurate, transparent, and flexible network alignments. open.substack.com/pub/bravoaba...
03.05.2025 14:31 — 👍 2 🔁 1 💬 0 📌 0Probabilistic alignment of multiple networks
->Nature | More info from EcoSearch