Postdoctoral position in the team of Alessandro Barducci in the CBS, focusing on multiscale simulations of RNA-driven condensates, with applications in synthetic biology.
More information :
www.cbs.cnrs.fr/images/jobs/...
The position is expected to start in early 2026, with some flexibility.
05.11.2025 17:43 — 👍 13 🔁 13 💬 1 📌 4
Thanks to my coauthors; Midas Segers, Andrea Parmeggiani and @ecarlon.bsky.social for this nice collaboration.
30.04.2025 22:56 — 👍 1 🔁 0 💬 0 📌 0
Taken together, our results support a scenario where interphase chromatin self-organizes into alternating α/β domains.
This spatial arrangement likely reflects the combined action of passive mechanisms (e.g. bridging interactions) and active loop extrusion processes.
30.04.2025 22:54 — 👍 1 🔁 0 💬 1 📌 0
This memory effect is due to looped domains which introduce periodic trajectories of the chromatin organization. The auxin treatment of cells inhibes actives processes of loop extrusion due to cohesin which leads to the disorganization of the beta phase and thus of the memory effect.
30.04.2025 22:53 — 👍 1 🔁 0 💬 1 📌 0
We further analyze non-Markovian effects by computing conditional distance distributions and three-point correlations.
These analyses reveal that local chromatin structure influences distal contacts over hundreds of kilobases, with stronger memory effects in WT cells compared to Auxin-treated ones.
30.04.2025 22:48 — 👍 1 🔁 0 💬 1 📌 0
To interpret the observed scaling and spatial heterogeneity, we introduce a heterogeneous random walk model.
Despite its simplicity, it reproduces key features of the data, including the transition between intra-loop and inter-loop regimes, and the emergent microphase-separated structure
30.04.2025 22:47 — 👍 1 🔁 0 💬 1 📌 0
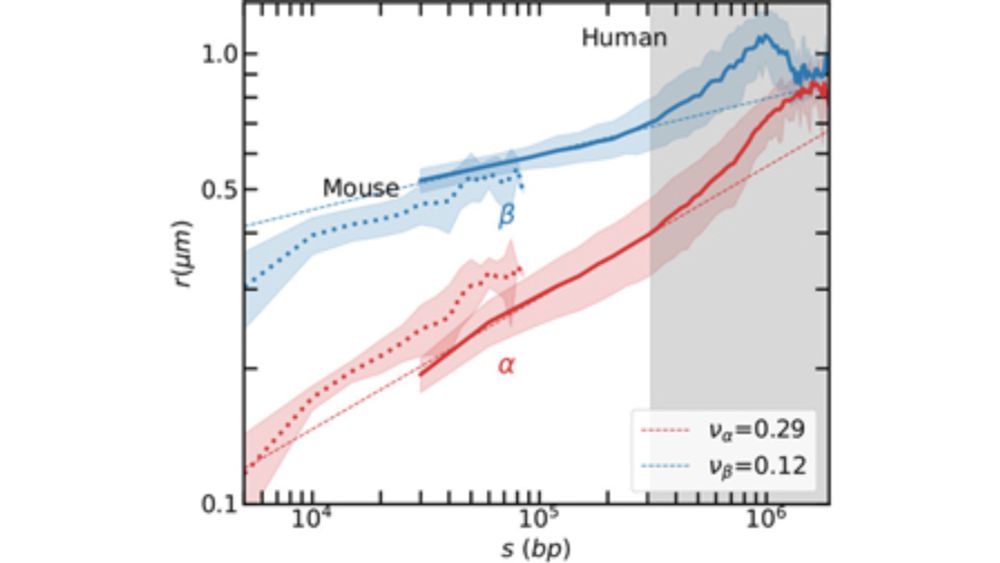
The α phase is consistent with a crumpled globule, a metastable polymer state which is spatially compact.
The β phase has a weaker exponent, suggesting a looped organization, possibly in the form of rosette-like domains.
This interpretation is supported by experimental data and analytical modeling.
30.04.2025 22:46 — 👍 1 🔁 0 💬 1 📌 0
The scaling behavior of these phases is analyzed in detail.
We find consistent results across human and mouse data, with distinct exponents for the α and β phases.
This allows us to characterize different regimes of chromatin folding across multiple levels of genome organization.
30.04.2025 22:41 — 👍 1 🔁 0 💬 1 📌 0
We focus on the probability distributions of pairwise spatial distances between labeled genomic loci.
These distributions reveal a robust two-component structure, well described by a superposition of Gaussians.
This statistical signature points to the coexistence of two conformations α and β phases
30.04.2025 22:36 — 👍 1 🔁 0 💬 1 📌 0
We analyze high-resolution multiplexed FISH (m-FISH) data from human and mouse cells, covering genomic scales from 5 kb to 2 Mb.
This allows us to compare and unify chromatin structural features across species and over a wide range of genomic distances.
30.04.2025 22:34 — 👍 1 🔁 0 💬 1 📌 0
Inferring interphase chromosomal structure from multiplexed fluorescence in situ hybridization data: A unified picture from human and mouse cells
We analyze multiplexed fluorescence in situ hybridization (m-FISH) data for human and mouse cell lines. The m-FISH technique uses fluorescently-labeled single-s
We have recently published the following article in The Journal of Chemical Physics:
“Inferring interphase chromosomal structure from multiplexed FISH data: a unified picture from human and mouse cells”
In this thread, I summarize the main findings and ideas discussed in the paper.
30.04.2025 22:33 — 👍 2 🔁 0 💬 1 📌 0
Congratulations @ecarlon.bsky.social !!!
28.04.2025 15:00 — 👍 0 🔁 0 💬 0 📌 0
I maxed out my credit card, emptied my savings account, and took out a loan to move from Alabama to Bethesda, MD. I don’t even qualify for unemployment since I’ve only worked at NIH for a month. I will be financially and medically devastated.
Seeking suggestions for anywhere that’s hiring!!
14.02.2025 23:49 — 👍 11548 🔁 4082 💬 671 📌 162
C'était hier soir
06.02.2025 12:16 — 👍 0 🔁 0 💬 0 📌 0
incroyable
06.02.2025 12:05 — 👍 0 🔁 0 💬 0 📌 0
The Swiss National Science Foundation funds excellent research at universities and other institutions – from chemistry to medicine to sociology.
We invest in researchers and their ideas 🔬🌱📚 — www.snf.ch/en
Deutsch & Français: @snf-fns.ch
Group Leader @EMBL. We study the design principles of "early" animals using cnidarians.
Optical tweezers | AFM | correlated fluorescence and force | nucleic acid-protein interactions | SSBs | chromatin stability | nucleosome chaperones | retrovirus, coronavirus, retrotransposon replication
https://williamslab.sites.northeastern.edu
Microbiologist | Biochemist | Structural Biologist, Lausanne 🇨🇭 | Studying molecular machines (SMC, ParB, Wadjet, Lamassu & more) in Genome Integrity & Nucleic Acid Immunity.
🔬 Lab homepage: wp.unil.ch/gruberlab
A career network featuring science jobs in academia and industry.
Visit our platform at www.science.hr
PhD student at CBS Montpellier 🇫🇷
MD simulations of proteins, RNA and condensates 🧬💻
News zum SNF, zu SNF-geförderter Forschung sowie zu nationaler & internationaler Forschungspolitik.
Actualités du FNS, des recherches qu’il finance et de la politique de recherche sur le plan national et international.
English: @snsf.ch
ICREA Research Professor at Universitat de Barcelona. Research Group Leader at MPI-PKS and CSBD in Dresden. Theory of living matter. Collective phenomena in biology through the lens of active matter physics.
Postdoc at the University of Geneva, specialising in structural color pigment formation in zebrafish using biophysical techniques and microscopy. Former PhD researcher @institutcurie.bsky.social, focused on cell adhesion on fluid substrates.
PostDoc, Kruse lab at UNIGE, working on active living matter and tissue mechanics.
Former PhD at Curie institute on cell & tissue mechanics.
IZTECH-Chemistry, I like to be Polymath(learning different subjects), Polygot(interested in the learning different languages) comments and opinions are my own RT≠ is not endorsement (he/him)
Science made simple. Sharp insights, byte by byte |Animals | Science |Technology |Plants |Health | Many More
📚 We're your source for papers on various #EpithelialMechanics topics 🔍 & platform to share your research and passion 🧫🔬
Like to write a thread? Please DM us!
💬 @onenimesa.bsky.social & @juliaeckert.bsky.social
👉 https://epithelialmechanics.github.io
Advancing the physical sciences with a unifying voice of strength from diversity. https://aip.org
10th Biennial European Cell Mechanics Meeting
30Sep-3Oct 2025 @KU_Leuven
https://cellmech2025.org/
CM: @barrasa-fano.bsky.social & Gabriela Marinho
The Groupe de Recherche Approches Quantitatives du Vivant (GdR AQV CNRS 2108) aims to consolidate and promote links between communities at the biology/physics interface.
Associate Professor at MIT BE : http://ashansenlab.com
Interested in understanding the relationship between 3D genome structure and function
Asst Prof at University of California, Irvine.
Genetics, Genomics, Gene Regulation, Development. Views are my own.
https://www.kvonlab.org/