OmicsPred is an atlas of genetic scores for prediction of multi-omics data.
We're working closely with our PredictDB colleagues to bring you an integrated resource for multi-omic predictors
We're happy to announce that PrediXcan GTExV8 expression predictors are now annotated and openly available via OmicsPred! www.omicspred.org
More info www.omicspred.org/publication/...
16.06.2025 12:24 — 👍 13 🔁 6 💬 0 📌 0
OmicsPred is developed by @laurentgil33.bsky.social @yuxu.bsky.social @iamslambert.bsky.social @mikeinouye.bsky.social with support from @uniofcam.bsky.social, HDRUK, @ebi.embl.org and the Baker Institute. Many thanks also to @hakyim.bsky.social, Ruidong Xiang, many others! (8/N)
06.02.2025 11:09 — 👍 6 🔁 2 💬 0 📌 1
We encourage users to contribute newly developed multi-omic genetic scores, fresh evaluations of existing scores, and new association results from additional cohorts. Sharing helps the community maximize utility and accessibility of these scores! Submit your scores: forms.gle/KLZGbvurrftF... (7/N)
06.02.2025 11:09 — 👍 0 🔁 1 💬 1 📌 0
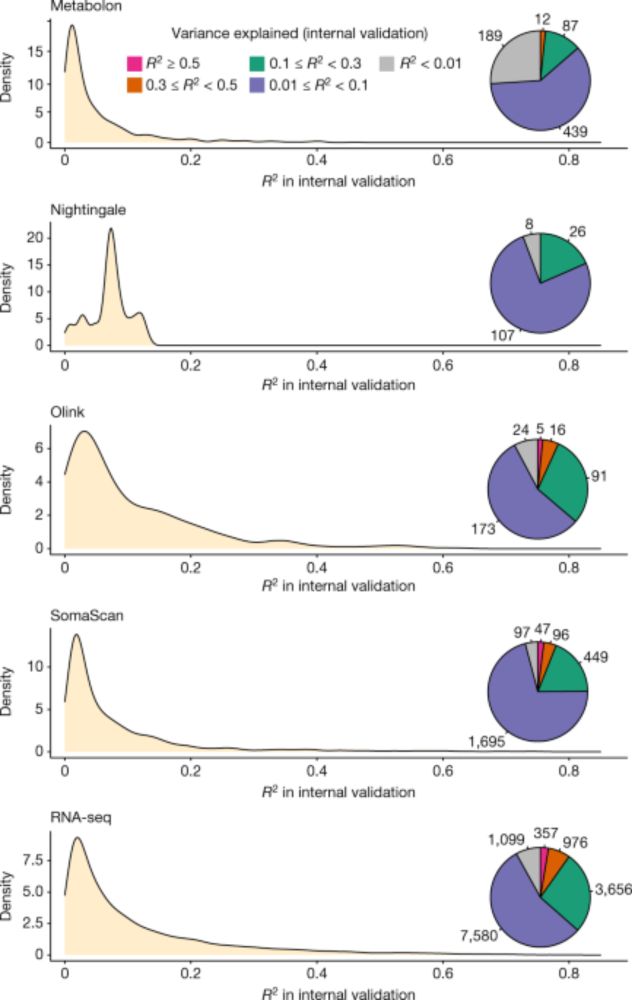
An atlas of genetic scores to predict multi-omic traits - Nature
A machine learning approach is used to analyse multi-omics (proteomics, metabolomics and transcriptomics) data, producing genetic scores for more than 17,000 biomolecular traits in human blood, a...
Currently, we've curated scores from our flagship paper (doi.org/10.1038/s415...), along with recently developed scores for ~3000 Olink proteomic traits from the UKB PPP (preprint incoming). We are collaborating with & curating cognate resources in the community for our next update. Stay tuned!(6/N)
06.02.2025 11:09 — 👍 3 🔁 1 💬 1 📌 0
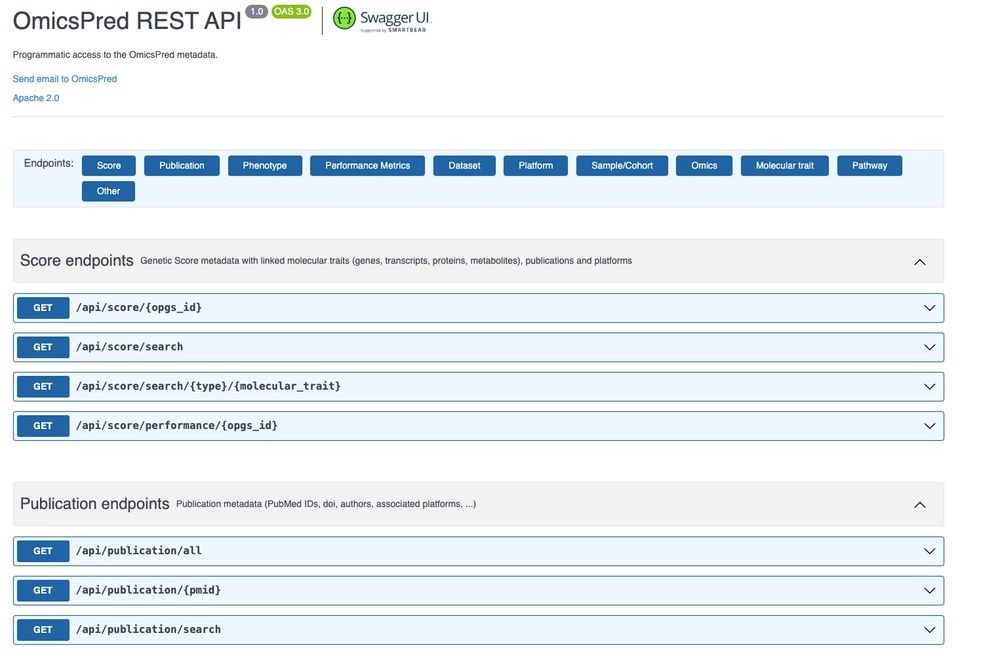
We provide REST APIs to allow users to search and retrieve all OmicsPred metadata, offering flexibility to identify scores, associations, or other entities that meet their specific needs. (5/N)
06.02.2025 11:09 — 👍 0 🔁 1 💬 1 📌 0
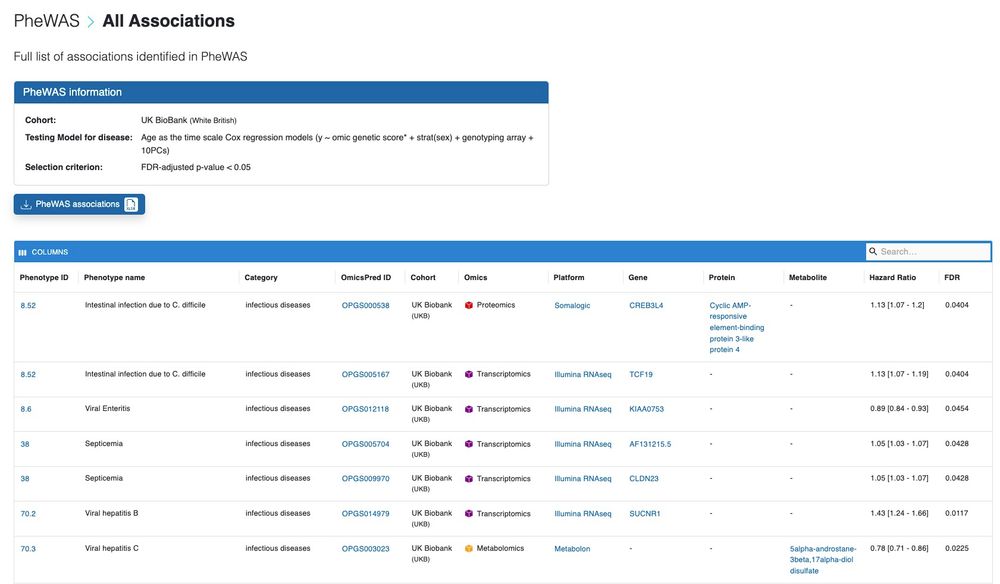
You can also browse and search for omic trait-phenotype associations identified in the biobank association analyses using the genetically predicted multi-omic traits. (4/N)
06.02.2025 11:09 — 👍 0 🔁 1 💬 1 📌 0
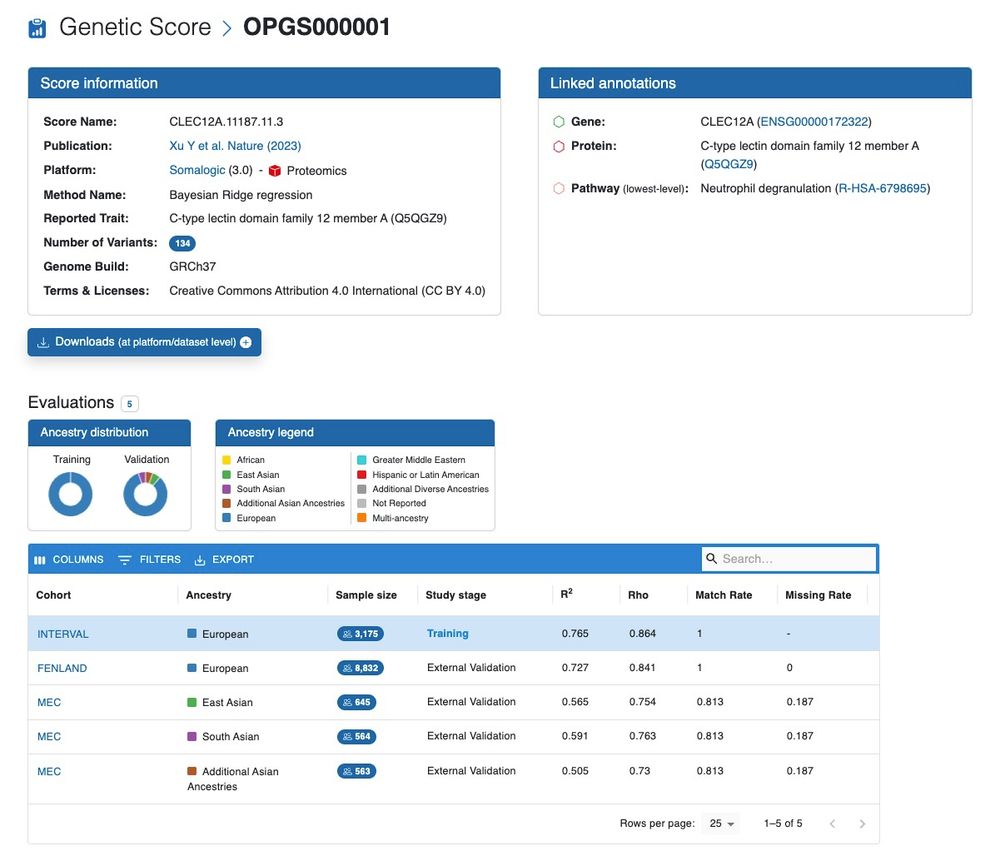
Each score has its own page describing the omic trait a score predicts, its linked biological annotations, how the score was developed, samples used for training/testing (with ancestry information) and related performances, and a link to download the score model. (3/N)
06.02.2025 11:09 — 👍 0 🔁 1 💬 1 📌 0
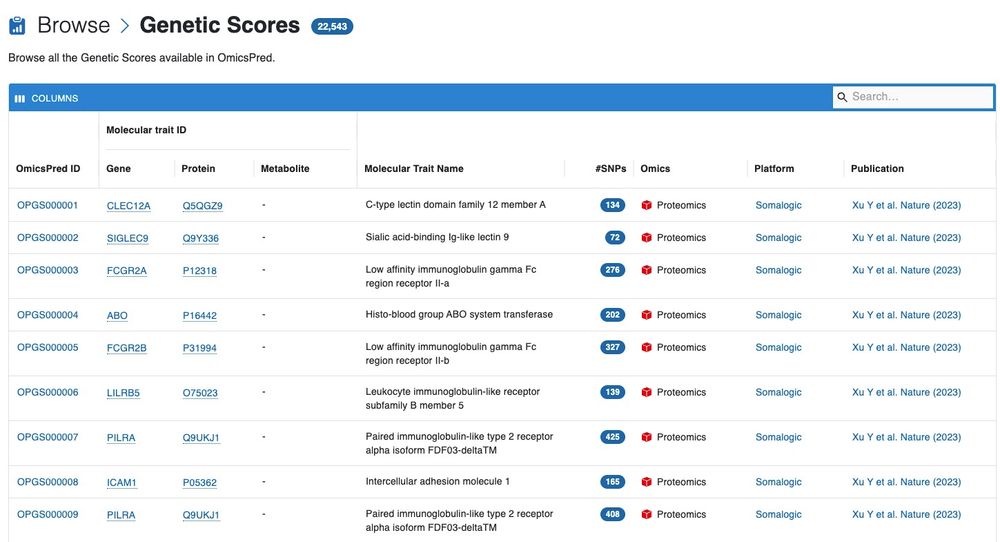
On OmicsPred, you can browse and search for multi-omic genetic scores (www.omicspred.org/scores), related publications, omics platforms, and the biological pathways associated with these scores. (2/N)
06.02.2025 11:09 — 👍 0 🔁 1 💬 1 📌 0
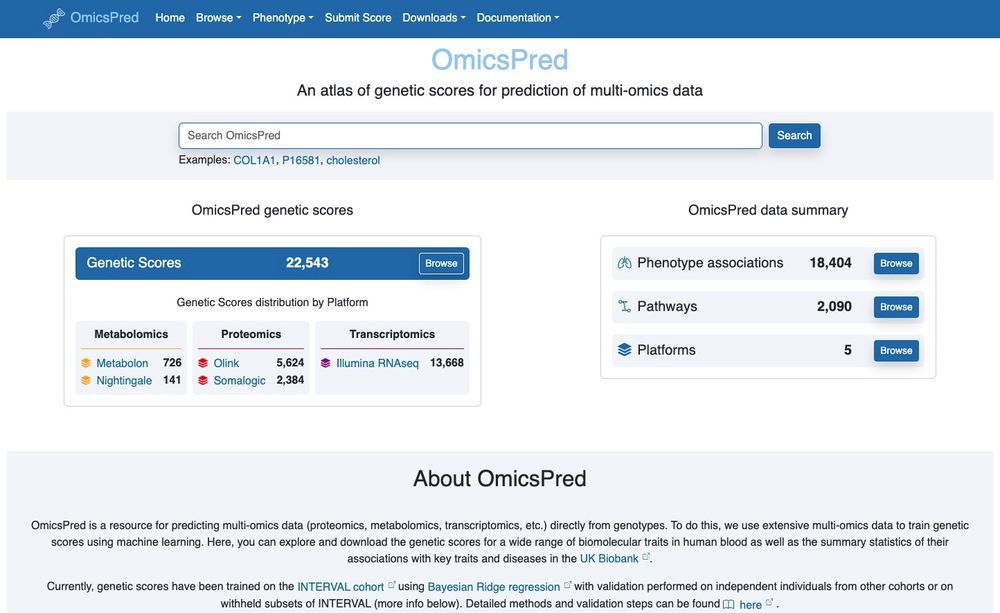
Introducing a major upgrade to OmicsPred platform (www.omicspred.org) — a resource to enhance the accessibility and usability of genetic scores for multi-omic traits and their phenotypic associations. (1/N)
06.02.2025 11:09 — 👍 15 🔁 13 💬 1 📌 2
A joint research initiative at the University of Cambridge and the Baker Heart & Diabetes Institute
Cambridge, UK | Melbourne, Australia
www.inouyelab.org
Senior Research Associate at Cambridge University
Genetic prediction, Multi-omics, Health data science, Machine Learning
🇨🇦 in 🇬🇧. Assistant Prof. studying polygenic scores & multimorbidity at University of Cambridge; co-lead @pgscatalog.bsky.social. Otherwise, probably talking about good films, bad tv, or wine.
Principal Software developer at Wellcome Sanger Institute
Current projects:
- OmicsPred: @omicspred.bsky.social
- PGS Catalog: @pgscatalog.bsky.social
Computational biologist. Geriatric Millennial. Professor, University of Cambridge. Director of Data Sciences, Baker Heart & Diabetes Institute. British | Australian | American.
www.inouyelab.org | Cambridge, UK
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app





